
Journal of Shanghai Jiao Tong University (Medical Science) ›› 2025, Vol. 45 ›› Issue (4): 387-403.doi: 10.3969/j.issn.1674-8115.2025.04.001
• Basic research • Next Articles
LI Linying1, CAI Xiaodong2, TONG Ran2, YANG Chen2, WANG Zhiming2, HE Xiaoyu2, MA Ziyue2, ZHANG Feng2( ), LI Lingjie2(
), LI Lingjie2( ), ZHOU Junmei1(
), ZHOU Junmei1( )
)
Received:2024-09-02
Accepted:2024-11-11
Online:2025-04-28
Published:2025-04-28
Contact:
ZHANG Feng, LI Lingjie, ZHOU Junmei
E-mail:fzhang@shsmu.edu.cn;lingjie@shsmu.edu.cn;zhou_junmei@qq.com
Supported by:CLC Number:
LI Linying, CAI Xiaodong, TONG Ran, YANG Chen, WANG Zhiming, HE Xiaoyu, MA Ziyue, ZHANG Feng, LI Lingjie, ZHOU Junmei. Analysis of transcriptome and chromatin accessibility changes during the differentiation of human embryonic stem cells into neural progenitor cells[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(4): 387-403.
Add to citation manager EndNote|Ris|BibTeX
URL: https://xuebao.shsmu.edu.cn/EN/10.3969/j.issn.1674-8115.2025.04.001
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| GAPDH | CTGAGAACGGGAAGCTTGT | GGGTGCTAAGCAGTTGGT |
| NANOG | GAATGAAATCTAAGAGGTGGCA | CCTGGTGGTAGGAAGAGTAAAGG |
| POU5F1 | ACATCAAAGCTCTGCAGAAAGAACT | CTGAATACCTTCCCAAATAGAACCC |
| SOX1 | GGAATGGGAGGACAGGATTT | ACTTTTATTTCTCGGCCCGT |
| PAX6 | GCCTATGCAACCCCCAGT | TCACTTCCGGGAACTTGAAC |
| NES | TGCGGGCTACTGAAAAGTTC | AGGCTGAGGGACATCTTGAG |
Tab 1 Primer sequences for RT-qPCR
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| GAPDH | CTGAGAACGGGAAGCTTGT | GGGTGCTAAGCAGTTGGT |
| NANOG | GAATGAAATCTAAGAGGTGGCA | CCTGGTGGTAGGAAGAGTAAAGG |
| POU5F1 | ACATCAAAGCTCTGCAGAAAGAACT | CTGAATACCTTCCCAAATAGAACCC |
| SOX1 | GGAATGGGAGGACAGGATTT | ACTTTTATTTCTCGGCCCGT |
| PAX6 | GCCTATGCAACCCCCAGT | TCACTTCCGGGAACTTGAAC |
| NES | TGCGGGCTACTGAAAAGTTC | AGGCTGAGGGACATCTTGAG |
| Index | Sample | |||
|---|---|---|---|---|
| ESC_S1 | ESC_S2 | NPC_S1 | NPC_S2 | |
| Total raw read pairs/n | 45 792 974 | 53 251 136 | 50 419 620 | 52 973 024 |
| Total raw bases/n | 6 914 739 074 | 8 040 921 536 | 7 613 362 620 | 7 998 926 624 |
| Mapped reads/n | 44 506 927 | 51 742 966 | 48 948 795 | 51 574 332 |
| Mapped ratio/% | 97.95 | 97.92 | 97.87 | 98.13 |
| High-quality mapped reads/n | 43 139 843 | 50 148 322 | 47 576 740 | 50 093 517 |
| High-quality mapped ratio/% | 94.94 | 94.90 | 95.13 | 95.31 |
Tab 2 Number of raw reads and mapped reads obtained from RNA-seq libraries during neural induction
| Index | Sample | |||
|---|---|---|---|---|
| ESC_S1 | ESC_S2 | NPC_S1 | NPC_S2 | |
| Total raw read pairs/n | 45 792 974 | 53 251 136 | 50 419 620 | 52 973 024 |
| Total raw bases/n | 6 914 739 074 | 8 040 921 536 | 7 613 362 620 | 7 998 926 624 |
| Mapped reads/n | 44 506 927 | 51 742 966 | 48 948 795 | 51 574 332 |
| Mapped ratio/% | 97.95 | 97.92 | 97.87 | 98.13 |
| High-quality mapped reads/n | 43 139 843 | 50 148 322 | 47 576 740 | 50 093 517 |
| High-quality mapped ratio/% | 94.94 | 94.90 | 95.13 | 95.31 |
| Index | Sample | |||
|---|---|---|---|---|
| ESC_S1 | ESC_S2 | NPC_S1 | NPC_S2 | |
| Total raw read pairs/n | 63 713 180 | 32 001 236 | 88 523 604 | 61 924 682 |
| Total raw bases/n | 9 620 690 180 | 4 832 186 636 | 13 367 000 000 | 9 350 626 982 |
| Mapped read pairs/n | 61 240 240 | 30 684 142 | 84 944 932 | 59 439 388 |
| Mapped ratio/% | 98.36 | 98.52 | 98.24 | 97.98 |
| High-quality mapped read pairs/n | 54 624 252 | 27 152 448 | 75 936 202 | 53 201 562 |
| High-quality mapped ratio/% | 87.73 | 87.18 | 87.82 | 87.70 |
| Percent duplication/% | 23.70 | 20.59 | 22.18 | 29.05 |
| Non-duplicate read pairs/n | 41 677 046 | 21 562 952 | 59 092 184 | 37 745 230 |
Tab 3 Number of raw reads and mapped reads obtained by ATAC-seq libraries during neural induction
| Index | Sample | |||
|---|---|---|---|---|
| ESC_S1 | ESC_S2 | NPC_S1 | NPC_S2 | |
| Total raw read pairs/n | 63 713 180 | 32 001 236 | 88 523 604 | 61 924 682 |
| Total raw bases/n | 9 620 690 180 | 4 832 186 636 | 13 367 000 000 | 9 350 626 982 |
| Mapped read pairs/n | 61 240 240 | 30 684 142 | 84 944 932 | 59 439 388 |
| Mapped ratio/% | 98.36 | 98.52 | 98.24 | 97.98 |
| High-quality mapped read pairs/n | 54 624 252 | 27 152 448 | 75 936 202 | 53 201 562 |
| High-quality mapped ratio/% | 87.73 | 87.18 | 87.82 | 87.70 |
| Percent duplication/% | 23.70 | 20.59 | 22.18 | 29.05 |
| Non-duplicate read pairs/n | 41 677 046 | 21 562 952 | 59 092 184 | 37 745 230 |
| Motif | E value | Motif ID | Motif | E value | Motif ID |
|---|---|---|---|---|---|
 | 2.38×10-111 | DLX1 | 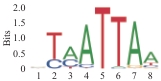 | 1.05×10-100 | NKX6.1 |
 | 6.26×10-109 | SHOX |  | 2.79×10-91 | LHX2 |
 | 6.18×10-101 | MSX1 |
Tab 4 Motif enrichment analysis of open chromatin signals that were differentially upregulated at the NPC stage
| Motif | E value | Motif ID | Motif | E value | Motif ID |
|---|---|---|---|---|---|
 | 2.38×10-111 | DLX1 |  | 1.05×10-100 | NKX6.1 |
 | 6.26×10-109 | SHOX |  | 2.79×10-91 | LHX2 |
 | 6.18×10-101 | MSX1 |
| Gene | Degree |
|---|---|
| EZH2 | 22 |
| PRKACA | 18 |
| PRKACB | 18 |
| JUN | 18 |
| FN1 | 16 |
| CDH2 | 15 |
| ERBB4 | 15 |
| RUNX1 | 15 |
| GSK3B | 14 |
| IFT88 | 13 |
| TUBA1A | 12 |
| PDGFRA | 11 |
| GNG2 | 11 |
| DYNC1I2 | 10 |
| H2BC21 | 10 |
| PIK3CA | 10 |
| PIK3R1 | 10 |
| MLLT3 | 10 |
Tab 5 Candidate downstream hub genes with the degrees≥10 in the PPI network
| Gene | Degree |
|---|---|
| EZH2 | 22 |
| PRKACA | 18 |
| PRKACB | 18 |
| JUN | 18 |
| FN1 | 16 |
| CDH2 | 15 |
| ERBB4 | 15 |
| RUNX1 | 15 |
| GSK3B | 14 |
| IFT88 | 13 |
| TUBA1A | 12 |
| PDGFRA | 11 |
| GNG2 | 11 |
| DYNC1I2 | 10 |
| H2BC21 | 10 |
| PIK3CA | 10 |
| PIK3R1 | 10 |
| MLLT3 | 10 |
| 1 | UCHIDA N, BUCK D W, HE D, et al. Direct isolation of human central nervous system stem cells[J]. Proc Natl Acad Sci USA, 2000, 97(26): 14720-14725. |
| 2 | NOCTOR S C, FLINT A C, WEISSMAN T A, et al. Neurons derived from radial glial cells establish radial units in neocortex[J]. Nature, 2001, 409(6821): 714-720. |
| 3 | KRIEGSTEIN A, ALVAREZ-BUYLLA A. The glial nature of embryonic and adult neural stem cells[J]. Annu Rev Neurosci, 2009, 32: 149-184. |
| 4 | MARCHETTO M C, BELINSON H, TIAN Y, et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals[J]. Mol Psychiatry, 2017, 22(6): 820-835. |
| 5 | HOWELL B W, SMITH K M. Synaptic structural protein dysfunction leads to altered excitation inhibition ratios in models of autism spectrum disorder[J]. Pharmacol Res, 2019, 139: 207-214. |
| 6 | BOZZI Y, PROVENZANO G, CASAROSA S. Neurobiological bases of autism-epilepsy comorbidity: a focus on excitation/inhibition imbalance[J]. Eur J Neurosci, 2018, 47(6): 534-548. |
| 7 | OLIVEIRA B, MITJANS M, NITSCHE M A, et al. Excitation-inhibition dysbalance as predictor of autistic phenotypes[J]. J Psychiatr Res, 2018, 104: 96-99. |
| 8 | MOOSAVI A, MOTEVALIZADEH ARDEKANI A. Role of epigenetics in biology and human diseases[J]. Iran Biomed J, 2016, 20(5): 246-258. |
| 9 | FITZ-JAMES M H, CAVALLI G. Molecular mechanisms of transgenerational epigenetic inheritance[J]. Nat Rev Genet, 2022, 23(6): 325-341. |
| 10 | ALLIS C D, JENUWEIN T. The molecular hallmarks of epigenetic control[J]. Nat Rev Genet, 2016, 17(8): 487-500. |
| 11 | DAI S K, LIU P P, LI X, et al. Dynamic profiling and functional interpretation of histone lysine crotonylation and lactylation during neural development[J]. Development, 2022, 149(14): dev200049. |
| 12 | SUN T Y, XU Y Y, XIANG Y, et al. Crosstalk between RNA m6A and DNA methylation regulates transposable element chromatin activation and cell fate in human pluripotent stem cells[J]. Nat Genet, 2023, 55(8): 1324-1335. |
| 13 | WANG C F, YANG J W, ZHUANG Z H, et al. Activity-dependent feedback regulation of thalamocortical axon development by Lhx2 in cortical layer 4 neurons[J]. Cereb Cortex, 2023, 33(5): 1693-1707. |
| 14 | TSENG C J, MCDOUGLE C J, HOOKER J M, et al. Epigenetics of autism spectrum disorder: histone deacetylases[J]. Biol Psychiatry, 2022, 91(11): 922-933. |
| 15 | CONBOY K, HENSHALL D C, BRENNAN G P. Epigenetic principles underlying epileptogenesis and epilepsy syndromes[J]. Neurobiol Dis, 2021, 148: 105179. |
| 16 | KLEMM S L, SHIPONY Z, GREENLEAF W J. Chromatin accessibility and the regulatory epigenome[J]. Nat Rev Genet, 2019, 20(4): 207-220. |
| 17 | SONG L Y, CRAWFORD G E. [J]. Cold Spring Harb Protoc, 2010, 2010(2): pdb.prot5384. |
| 18 | BUENROSTRO J D, GIRESI P G, ZABA L C, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position[J]. Nat Methods, 2013, 10(12): 1213-1218. |
| 19 | MARKENSCOFF-PAPADIMITRIOU E, WHALEN S, PRZYTYCKI P, et al. A chromatin accessibility atlas of the developing human telencephalon[J]. Cell, 2020, 182(3): 754-769.e18. |
| 20 | LU C Y, GARIPLER G, DAI C, et al. Essential transcription factors for induced neuron differentiation[J]. Nat Commun, 2023, 14(1): 8362. |
| 21 | THOMSON J A, ITSKOVITZ-ELDOR J, SHAPIRO S S, et al. Embryonic stem cell lines derived from human blastocysts[J]. Science, 1998, 282(5391): 1145-1147. |
| 22 | TAKAHASHI K, TANABE K, OHNUKI M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors[J]. Cell, 2007, 131(5): 861-872. |
| 23 | YU J Y, VODYANIK M A, SMUGA-OTTO K, et al. Induced pluripotent stem cell lines derived from human somatic cells[J]. Science, 2007, 318(5858): 1917-1920. |
| 24 | CHAMBERS S M, FASANO C A, PAPAPETROU E P, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling[J]. Nat Biotechnol, 2009, 27(3): 275-280. |
| 25 | KAWASAKI H, MIZUSEKI K, NISHIKAWA S, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity[J]. Neuron, 2000, 28(1): 31-40. |
| 26 | LEE H, SHAMY G A, ELKABETZ Y, et al. Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons[J]. Stem Cells, 2007, 25(8): 1931-1939. |
| 27 | ZHANG S C, WERNIG M, DUNCAN I D, et al. In vitro differentiation of transplantable neural precursors from human embryonic stem cells[J]. Nat Biotechnol, 2001, 19(12): 1129-1133. |
| 28 | TCHIEU J, ZIMMER B, FATTAHI F, et al. A modular platform for differentiation of human PSCs into all major ectodermal lineages[J]. Cell Stem Cell, 2017, 21(3): 399-410.e7. |
| 29 | MARTIN M. Cutadapt removes adapter sequences from high-throughput sequencing reads[J]. EMBnet J, 2011, 17(1): 10-12. |
| 30 | BOLGER A M, LOHSE M, USADEL B. Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30(15): 2114-2120. |
| 31 | PATEL R K, JAIN M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data[J]. PLoS One, 2012, 7(2): e30619. |
| 32 | KIM D, LANGMEAD B, SALZBERG S L. HISAT: a fast spliced aligner with low memory requirements[J]. Nat Methods, 2015, 12(4): 357-360. |
| 33 | LIAO Y, SMYTH G K, SHI W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features[J]. Bioinformatics, 2014, 30(7): 923-930. |
| 34 | LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. |
| 35 | YU G C, WANG L G, HAN Y Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16(5): 284-287. |
| 36 | LANGMEAD B, SALZBERG S L. Fast gapped-read alignment with bowtie 2[J]. Nat Methods, 2012, 9(4): 357-359. |
| 37 | ZHANG Y, LIU T, MEYER C A, et al. Model-based analysis of ChIP-seq (MACS)[J]. Genome Biol, 2008, 9(9): R137. |
| 38 | BAILEY T L, BODEN M, BUSKE F A, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| 39 | ROSS-INNES C S, STARK R, TESCHENDORFF A E, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer[J]. Nature, 2012, 481(7381): 389-393. |
| 40 | WATANABE K, UENO M, KAMIYA D, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells[J]. Nat Biotechnol, 2007, 25(6): 681-686. |
| 41 | ITSKOVITZ-ELDOR J, SCHULDINER M, KARSENTI D, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers[J]. Mol Med, 2000, 6(2): 88-95. |
| 42 | SZKLARCZYK D, KIRSCH R, KOUTROULI M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest[J]. Nucleic Acids Res, 2023, 51(D1): D638-D646. |
| 43 | JIA E T, PAN M, LIU Z Y, et al. Transcriptomic profiling of differentially expressed genes and related pathways in different brain regions in Parkinson's disease[J]. Neurosci Lett, 2020, 732: 135074. |
| 44 | ARAYA C, HÄKKINEN H M, CARCAMO L, et al. Cdh2 coordinates Myosin-Ⅱ dependent internalisation of the zebrafish neural plate[J]. Sci Rep, 2019, 9(1): 1835. |
| 45 | BARETTINO C, BALLESTEROS-GONZALEZ Á, AYLÓN A, et al. Developmental disruption of Erbb4 in Pet1+ neurons impairs serotonergic sub-system connectivity and memory formation[J]. Front Cell Dev Biol, 2021, 9: 770458. |
| 46 | TUNCAY I O, PARMALEE N L, KHALIL R, et al. Analysis of recent shared ancestry in a familial cohort identifies coding and noncoding autism spectrum disorder variants[J]. NPJ Genom Med, 2022, 7(1): 13. |
| 47 | ASADOLLAHI R, ONEDA B, JOSET P, et al. The clinical significance of small copy number variants in neurodevelopmental disorders[J]. J Med Genet, 2014, 51(10): 677-688. |
| 48 | CHEN Z N, LI X J, CUI X Z, et al. Association of CTNND2 gene polymorphism with schizophrenia: two-sample case-control study in Chinese Han population[J]. Int J Psychiatry Med, 2023, 58(5): 433-448. |
| 49 | WANG L Y, XU M, WANG Y, et al. Melatonin improves synapse development by PI3K/Akt signaling in a mouse model of autism spectrum disorder[J]. Neural Regen Res, 2024, 19(7): 1618-1624. |
| 50 | VAZ R, EDWARDS S, DUEÑAS-REY A, et al. Loss of ctnnd2b affects neuronal differentiation and behavior in zebrafish[J]. Front Neurosci, 2023, 17: 1205653. |
| 51 | SCHMID C M, GREGOR A, COSTAIN G, et al. LHX2 haploinsufficiency causes a variable neurodevelopmental disorder[J]. Genet Med, 2023, 25(7): 100839. |
| 52 | WANG Y N, KHANDELWAL N, LIU S Q, et al. KDM6B cooperates with Tau and regulates synaptic plasticity and cognition via inducing VGLUT1/2[J]. Mol Psychiatry, 2022, 27(12): 5213-5226. |
| 53 | LI W, SHEN W C, ZHANG B, et al. Long non-coding RNA LncKdm2b regulates cortical neuronal differentiation by cis-activating Kdm2b[J]. Protein Cell, 2020, 11(3): 161-186. |
| 54 | CHEN D H, MCMANUS C E, RADMANESH B, et al. Temporal inhibition of chromatin looping and enhancer accessibility during neuronal remodeling[J]. Nat Commun, 2021, 12(1): 6366. |
| 55 | RAHMAN S, DONG P F, APONTES P, et al. Lineage specific 3D genome structure in the adult human brain and neurodevelopmental changes in the chromatin interactome[J]. Nucleic Acids Res, 2023, 51(20): 11142-11161. |
| 56 | CORCES M R, SHCHERBINA A, KUNDU S, et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer's and Parkinson's diseases[J]. Nat Genet, 2020, 52(11): 1158-1168. |
| 57 | PAGNI S, MILLS J D, FRANKISH A, et al. Non-coding regulatory elements: potential roles in disease and the case of epilepsy[J]. Neuropathol Appl Neurobiol, 2022, 48(3): e12775. |
| [1] | XU Wenhui, YANG Chang, LI Ruiqing, BIAN Jing, LI Xiayi, ZHENG Leizhen. Exploratory study of interferon regulatory factor 3 promoting proliferation and invasion related to colorectal cancer cells [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(3): 301-311. |
| [2] | ZHOU Yue, CHENG Chen, ZHENG Enlin, MENG Zhuo, WANG Jian, WANG Qingjie, HE Yongning, SUN Kun. Exploring potential new receptors for ELABELA in human embryonic stem cells by Crispr/Cas9-mediated gene editing system [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2022, 42(9): 1258-1264. |
| [3] | ZHAO Bingnan, MA Shuangyu, XU Chunlong, WANG Qiong. Different roles of SMAD2 and SMAD3 in human embryonic stem cell differentiation [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2022, 42(4): 443-454. |
| [4] | Xiao-zhi SUN, Shuang LI, Ying JIN, Bing LIAO. Effect of enzyme digestion into cell clumps on protein levels of OCT4 and SOX2 in human embryonic stem cells [J]. JOURNAL OF SHANGHAI JIAOTONG UNIVERSITY (MEDICAL SCIENCE), 2021, 41(4): 413-420. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||