
Journal of Shanghai Jiao Tong University (Medical Science) ›› 2023, Vol. 43 ›› Issue (5): 545-559.doi: 10.3969/j.issn.1674-8115.2023.05.005
• Innovative research team achievement column • Previous Articles Next Articles
MEI Yanqing1( ), HAN Yujie1, WENG Wenyun1, ZHANG Lei1,2, TANG Yujie1,2(
), HAN Yujie1, WENG Wenyun1, ZHANG Lei1,2, TANG Yujie1,2( )
)
Received:2023-03-08
Accepted:2023-04-10
Online:2023-05-28
Published:2023-07-11
Contact:
TANG Yujie
E-mail:yanqingmei@sjtu.edu.cn;yujietang@shsmu.edu.cn
Supported by:CLC Number:
MEI Yanqing, HAN Yujie, WENG Wenyun, ZHANG Lei, TANG Yujie. In vitro therapeutic effects and molecular mechanisms of targeted inhibition of CDK12/13 in high-grade gliomas[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(5): 545-559.
Add to citation manager EndNote|Ris|BibTeX
URL: https://xuebao.shsmu.edu.cn/EN/10.3969/j.issn.1674-8115.2023.05.005
| Primer | Sequence (5'→3') |
|---|---|
| sgGFP forward | CACCGGGGCGAGGAGCTGTTCACCG |
| sgGFP reverse | AAACCGGTGAACAGCTCCTCGCCCC |
| sgCDK12-2 forward | CACCGACTGACCGACTGCCTTCT |
| sgCDK12-2 reverse | AAACCGAGAAGGCAGTCGGTCAGTC |
| sgCDK12-3 forward | CACCGATTCACCAGTTCAGTATCTG |
| sgCDK12-3 reverse | AAACCAGATACTGAACTGGTGAATC |
Tab 1 Oligo sequences of sgRNA
| Primer | Sequence (5'→3') |
|---|---|
| sgGFP forward | CACCGGGGCGAGGAGCTGTTCACCG |
| sgGFP reverse | AAACCGGTGAACAGCTCCTCGCCCC |
| sgCDK12-2 forward | CACCGACTGACCGACTGCCTTCT |
| sgCDK12-2 reverse | AAACCGAGAAGGCAGTCGGTCAGTC |
| sgCDK12-3 forward | CACCGATTCACCAGTTCAGTATCTG |
| sgCDK12-3 reverse | AAACCAGATACTGAACTGGTGAATC |
| Primer | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| ATM | GCTGACAATCATCACCAAGT | GGTTCTCAGCACTATGGGACA |
| ATR | CGCTGAACTGTACGTGGAAA | CAATTAGTGCCTGGTGAACATC |
| BRCA1 | CTGCTCAGGGCTATCCTCTCA | GCTTCTAGTTCAGCCATTTCCTG |
| CCNK | AAAGCCATGTTGGTACTGGGA | TGTAGCCCCAAACGTGTGC |
| E2F1 | CATCAGTACCTGGCCGAGAG | CCCGGGGATTTCACACCTTT |
| FANCD2 | CCCAGAACTGATCAACTCTCCT | CCATCATCACACGGAAGAAA |
| FANCI | CACCACACTTACAGCCCTTG | ATTCCTCCGGAGCTCTGAC |
| RAD51 | GCTGATGAGTTTGGTGTAGCAG | GGAAGACAGGGAGAGTCGTAGA |
| SMARCC2 | GAGGGATAAGCAGGTTCTTCTG | GGGATCCACGTGTCGTAACT |
| TP53 | CCCAAGCAATGGATGATTTGA | GCATTCTGGGAGCTTCATCT |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA |
| dGAPDH | CGTTCATGCCACCACCGCTA | CCACGTCCATCACGCCACAA |
Tab 2 Primer sequence for RT-qPCR
| Primer | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| ATM | GCTGACAATCATCACCAAGT | GGTTCTCAGCACTATGGGACA |
| ATR | CGCTGAACTGTACGTGGAAA | CAATTAGTGCCTGGTGAACATC |
| BRCA1 | CTGCTCAGGGCTATCCTCTCA | GCTTCTAGTTCAGCCATTTCCTG |
| CCNK | AAAGCCATGTTGGTACTGGGA | TGTAGCCCCAAACGTGTGC |
| E2F1 | CATCAGTACCTGGCCGAGAG | CCCGGGGATTTCACACCTTT |
| FANCD2 | CCCAGAACTGATCAACTCTCCT | CCATCATCACACGGAAGAAA |
| FANCI | CACCACACTTACAGCCCTTG | ATTCCTCCGGAGCTCTGAC |
| RAD51 | GCTGATGAGTTTGGTGTAGCAG | GGAAGACAGGGAGAGTCGTAGA |
| SMARCC2 | GAGGGATAAGCAGGTTCTTCTG | GGGATCCACGTGTCGTAACT |
| TP53 | CCCAAGCAATGGATGATTTGA | GCATTCTGGGAGCTTCATCT |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA |
| dGAPDH | CGTTCATGCCACCACCGCTA | CCACGTCCATCACGCCACAA |
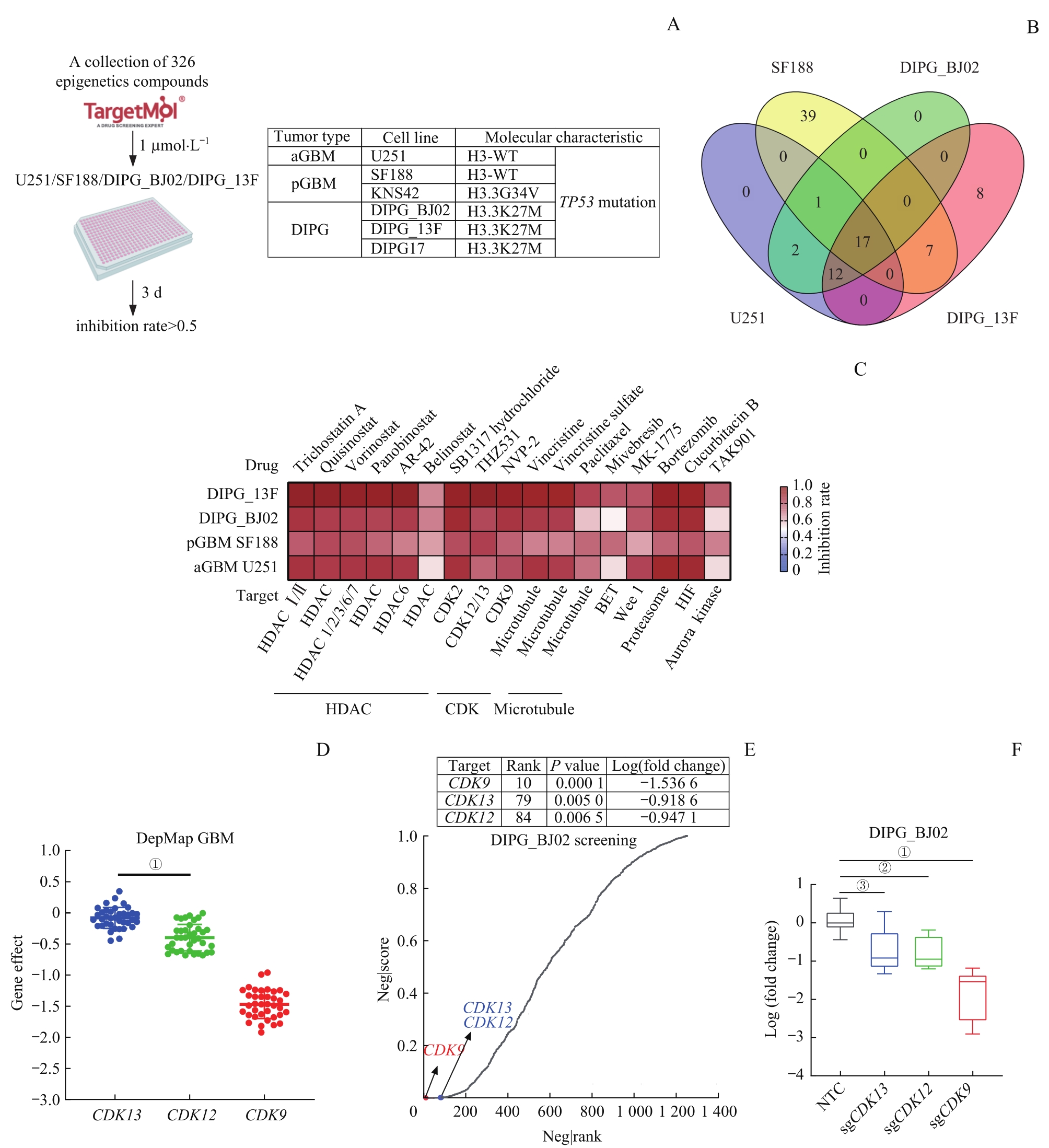
Fig 1 Screening for common potential drug targets for GBM and DIPG based on epigenetic transcription-related targeted small molecule drug library and functional genome: CDK12 and CDK13
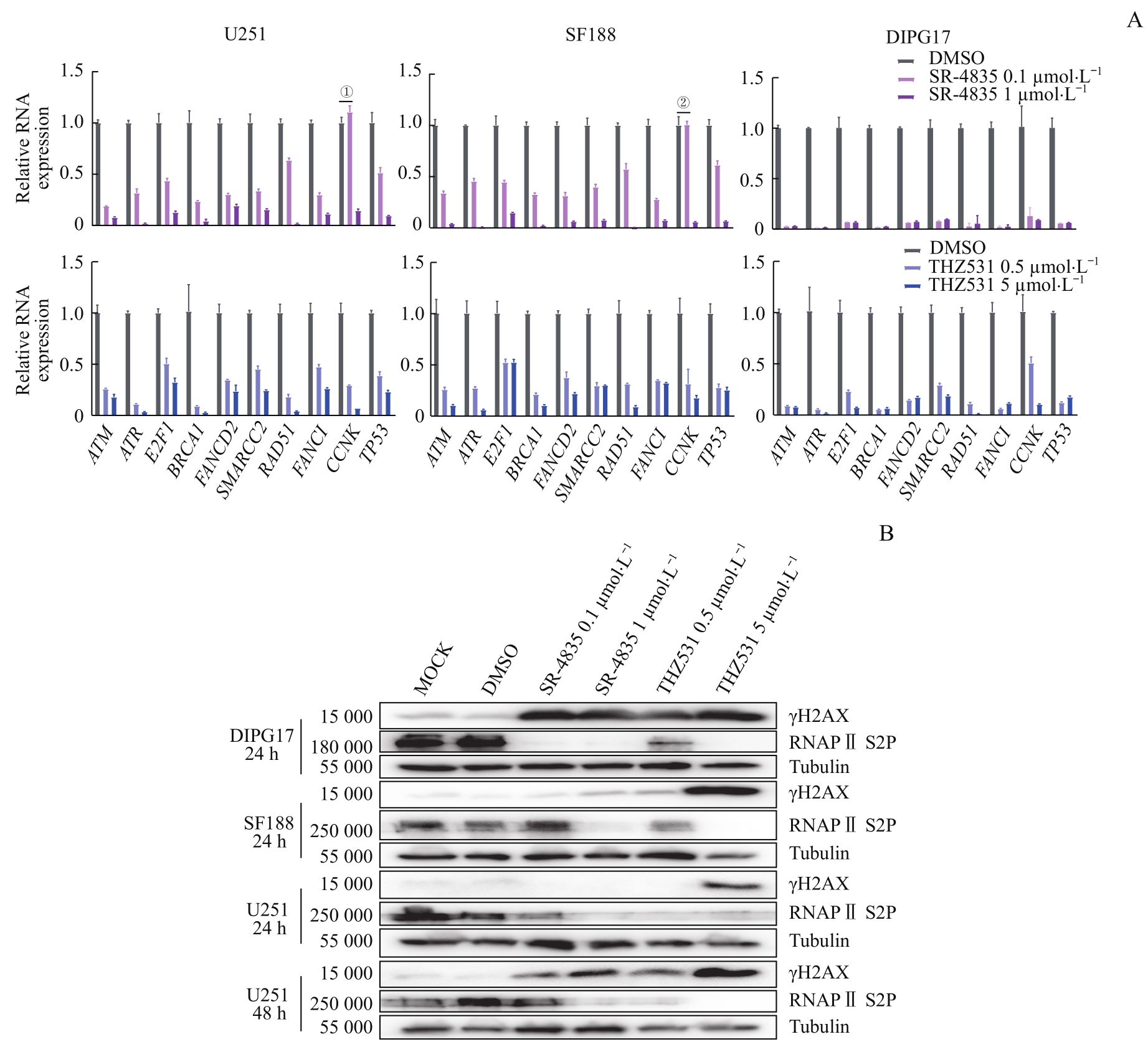
Fig 5 Targeted inhibition of CDK12/13 induced accumulation of DNA damage by significantly down-regulating transcription of genes involved in DDR in GBM and DIPG cells
| 1 | MILLER K D, OSTROM Q T, KRUCHKO C, et al. Brain and other central nervous system tumor statistics, 2021[J]. CA Cancer J Clin, 2021, 71(5): 381-406. |
| 2 | CHOI S, YU Y, GRIMMER M R, et al. Temozolomide-associated hypermutation in gliomas[J]. Neuro Oncol, 2018, 20(10): 1300-1309. |
| 3 | OSTROM Q T, PRICE M, NEFF C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015—2019[J]. Neuro Oncol, 2022, 24(Suppl 5): v1-v95. |
| 4 | MCKINNON C, NANDHABALAN M, MURRAY S A, et al. Glioblastoma: clinical presentation, diagnosis, and management[J]. BMJ, 2021, 374: n1560. |
| 5 | BRENNAN C W, VERHAAK R G, MCKENNA A, et al. The somatic genomic landscape of glioblastoma[J]. Cell, 2013, 155(2): 462-477. |
| 6 | MENG W, WANG J J, WANG B C, et al. CDK7 inhibition is a novel therapeutic strategy against GBM both in vitro and in vivo[J]. Cancer Manag Res, 2018, 10: 5747-5758. |
| 7 | YAN G, WANG Y F, CHEN J C, et al. Advances in drug development for targeted therapies for glioblastoma[J]. Med Res Rev, 2020, 40(5): 1950-1972. |
| 8 | NJONKOU R, JACKSON C M, WOODWORTH G F, et al. Pediatric glioblastoma: mechanisms of immune evasion and potential therapeutic opportunities[J]. Cancer Immunol Immunother, 2022, 71(8): 1813-1822. |
| 9 | BERGER T R, WEN P Y, LANG-ORSINI M, et al. World Health Organization 2021 classification of central nervous system tumors and implications for therapy for adult-type gliomas: a review[J]. JAMA Oncol, 2022, 8(10): 1493-1501. |
| 10 | ŚLEDZIŃSKA P, BEBYN M G, FURTAK J, et al. Prognostic and predictive biomarkers in gliomas[J]. Int J Mol Sci, 2021, 22(19): 10373. |
| 11 | PATHANIA M, DE JAY N, MAESTRO N, et al. H3.3K27M cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas[J]. Cancer Cell, 2017, 32(5): 684-700.e9. |
| 12 | HOFFMAN L M, VELDHUIJZEN VAN ZANTEN S E M, COLDITZ N, et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): a collaborative report from the international and European society for pediatric oncology DIPG registries[J]. J Clin Oncol, 2018, 36(19): 1963-1972. |
| 13 | AZIZ-BOSE R, MONJE M. Diffuse intrinsic pontine glioma: molecular landscape and emerging therapeutic targets[J]. Curr Opin Oncol, 2019, 31(6): 522-530. |
| 14 | RASHED W M, MAHER E, ADEL M, et al. Pediatric diffuse intrinsic pontine glioma: where do we stand?[J]. Cancer Metastasis Rev, 2019, 38(4): 759-770. |
| 15 | COONEY T M, LUBANSZKY E, PRASAD R, et al. Diffuse midline glioma: review of epigenetics[J]. J Neurooncol, 2020, 150(1): 27-34. |
| 16 | MENG W, WANG B C, MAO W W, et al. Enhanced efficacy of histone deacetylase inhibitor combined with bromodomain inhibitor in glioblastoma[J]. J Exp Clin Cancer Res, 2018, 37(1): 241. |
| 17 | MO J L, TAN K Z, DONG Y, et al. Therapeutic targeting the oncogenic driver EWSR1-FLI1 in Ewing sarcoma through inhibition of the FACT complex[J]. Oncogene, 2023, 42(1): 11-25. |
| 18 | NAGARAJA S, VITANZA N A, WOO P J, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma[J]. Cancer Cell, 2017, 31(5): 635-652.e6. |
| 19 | NGUYEN T T T, ZHANG Y R, SHANG E Y, et al. HDAC inhibitors elicit metabolic reprogramming by targeting super-enhancers in glioblastoma models[J]. J Clin Invest, 2020, 130(7): 3699-3716. |
| 20 | ANASTAS J N, ZEE B M, KALIN J H, et al. Re-programing chromatin with a bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG[J]. Cancer Cell, 2019, 36(5): 528-544.e10. |
| 21 | DAHL N A, DANIS E, BALAKRISHNAN I, et al. Super elongation complex as a targetable dependency in diffuse midline glioma[J]. Cell Rep, 2020, 31(1): 107485. |
| 22 | RANJAN A, PANG Y, BUTLER M, et al. Targeting CDK9 for the treatment of glioblastoma[J]. Cancers (Basel), 2021, 13(12): 3039. |
| 23 | CHOU J, QUIGLEY D A, ROBINSON T M, et al. Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy[J]. Cancer Discov, 2020, 10(3): 351-370. |
| 24 | LIANG K W, GAO X, GILMORE J M, et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing[J]. Mol Cell Biol, 2015, 35(6): 928-938. |
| 25 | CHOI S H, KIM S, JONES K A. Gene expression regulation by CDK12: a versatile kinase in cancer with functions beyond CTD phosphorylation[J]. Exp Mol Med, 2020, 52(5): 762-771. |
| 26 | QUEREDA V, BAYLE S, VENA F, et al. Therapeutic targeting of CDK12/CDK13 in triple-negative breast cancer[J]. Cancer Cell, 2019, 36(5): 545-558.e7. |
| 27 | WANG C, WANG H, LIEFTINK C, et al. CDK12 inhibition mediates DNA damage and is synergistic with sorafenib treatment in hepatocellular carcinoma[J]. Gut, 2020, 69(4): 727-736. |
| 28 | DIETER S M, SIEGL C, CODÓ P L, et al. Degradation of CCNK/CDK12 is a druggable vulnerability of colorectal cancer[J]. Cell Rep, 2021, 36(3): 109394. |
| 29 | LIU H, SHIN S H, CHEN H Y, et al. CDK12 and PAK2 as novel therapeutic targets for human gastric cancer[J]. Theranostics, 2020, 10(14): 6201-6215. |
| 30 | JIANG B S, JIANG J, KALTHEUNER I H, et al. Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma[J]. Eur J Med Chem, 2021, 221: 113481. |
| 31 | BLAZEK D, KOHOUTEK J, BARTHOLOMEEUSEN K, et al. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes[J]. Genes Dev, 2011, 25(20): 2158-2172. |
| 32 | DUBBURY S J, BOUTZ P L, SHARP P A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation[J]. Nature, 2018, 564(7734): 141-145. |
| 33 | ZHANG T H, KWIATKOWSKI N, OLSON C M, et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors[J]. Nat Chem Biol, 2016, 12(10): 876-884. |
| 34 | FENG J X, MEYER C A, WANG Q, et al. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data[J]. Bioinformatics, 2012, 28(21): 2782-2788. |
| 35 | INIGUEZ A B, STOLTE B, WANG E J, et al. EWS/FLI confers tumor cell synthetic lethality to CDK12 inhibition in Ewing sarcoma[J]. Cancer Cell, 2018, 33(2): 202-216.e6. |
| 36 | COLLINS P L, PURMAN C, PORTER S I, et al. DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner[J]. Nat Commun, 2020, 11(1): 3158. |
| 37 | LI R, OKADA H, YAMASHITA T, et al. FOXM1 is a novel molecular target of AFP-positive hepatocellular carcinoma abrogated by proteasome inhibition[J]. Int J Mol Sci, 2022, 23(15): 8305. |
| 38 | HOPKINS J L, ZOU L. Induction of BRCAness in triple-negative breast cancer by a CDK12/13 inhibitor improves chemotherapy[J]. Cancer Cell, 2019, 36(5): 461-463. |
| 39 | NIU T, LI K L, JIANG L, et al. Noncovalent CDK12/13 dual inhibitors-based PROTACs degrade CDK12-cyclin K complex and induce synthetic lethality with PARP inhibitor[J]. Eur J Med Chem, 2022, 228: 114012. |
| 40 | YAM C Q X, LIM H H, SURANA U. DNA damage checkpoint execution and the rules of its disengagement[J]. Front Cell Dev Biol, 2022, 10: 1020643. |
| 41 | XIE J B, SHEN Z Y, ANRAKU Y, et al. Nanomaterial-based blood-brain-barrier (BBB) crossing strategies[J]. Biomaterials, 2019, 224: 119491. |
| 42 | BOBO R H, LASKE D W, AKBASAK A, et al. Convection-enhanced delivery of macromolecules in the brain[J]. Proc Natl Acad Sci U S A, 1994, 91(6): 2076-2080. |
| 43 | MADANI F, LINDBERG S, LANGEL U, et al. Mechanisms of cellular uptake of cell-penetrating peptides[J]. J Biophys, 2011, 2011: 414729. |
| 44 | HERAVI SHARGH V, LUCKETT J, BOUZINAB K, et al. Chemosensitization of temozolomide-resistant pediatric diffuse midline glioma using potent nanoencapsulated forms of a N(3)-propargyl analogue[J]. ACS Appl Mater Interfaces, 2021, 13(30): 35266-35280. |
| 45 | WU T T, LIU Y, CAO Y, et al. Engineering macrophage exosome disguised biodegradable nanoplatform for enhanced sonodynamic therapy of glioblastoma[J]. Adv Mater, 2022, 34(15): e2110364. |
| [1] | Rui LI, Yu-jie HAN, Lei ZHANG, Yu-jie TANG. Invitro screening and validation of novel targeted therapeutic strategy against diffuse intrinsic pontine glioma [J]. JOURNAL OF SHANGHAI JIAOTONG UNIVERSITY (MEDICAL SCIENCE), 2021, 41(8): 987-998. |
| [2] | LI Chao, MI Jian-qing, WANG Jin. Advances in Philadelphia chromosome-like acute lymphoblastic leukemia [J]. JOURNAL OF SHANGHAI JIAOTONG UNIVERSITY (MEDICAL SCIENCE), 2020, 40(9): 1294-1301. |
| [3] | ZHANG Hui-lin, ZHU Xiao-na, YANG Shuo, LIU Meng-di, ZHU Di, YU Yun. Generation and phenotypic analysis of Fbxo22knockout mice [J]. , 2019, 39(4): 353-. |
| [4] | WENG Zhen1, NIU Xiao-yin2, ZHOU Jun-song1. Application of CRISPR-Cas9 technology in non-cancerous hematological disorders [J]. , 2018, 38(11): 1396-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||