
上海交通大学学报(医学版) ›› 2024, Vol. 44 ›› Issue (6): 663-675.doi: 10.3969/j.issn.1674-8115.2024.06.001
• 牙颌面畸形专题 • 下一篇
收稿日期:2024-02-29
接受日期:2024-04-01
出版日期:2024-06-28
发布日期:2024-06-28
通讯作者:
江凌勇
E-mail:jianglingyong@sjtu.edu.cn
作者简介:江凌勇(1978—),男,主任医师,博士;电子信箱:jianglingyong@sjtu.edu.cn。
基金资助:Received:2024-02-29
Accepted:2024-04-01
Online:2024-06-28
Published:2024-06-28
Contact:
JIANG Lingyong
E-mail:jianglingyong@sjtu.edu.cn
Supported by:摘要:
牙颌面骨畸形发病率高、病因复杂、症状严重、诊疗困难,缺乏早期干预策略,其主要原因在于相关机制研究较少、不够深入。这类疾病主要表现为骨性畸形、牙列不齐等骨与牙的形态结构、位置关系及口颌功能异常,其中颌面骨与牙-牙周复合体是两大核心结构,分别决定了颜面美观与咬合功能。颌面骨畸形精准防治需从病因角度研究发育与致病机制,而牙列不齐等牙-牙周复合体畸形矫治则需从临床正畸应力角度研究稳态与应激改建机制,两方面机制研究均可为牙颌面骨畸形防治策略发展提供重要理论基础。既往相关研究常以突变基因与差异因子表达谱的描述为主。近年来,Cre-LoxP等条件性基因编辑技术的发展,使研究者得以在体内直观地评价单一细胞谱系中致病基因的功能,助力牙颌面骨畸形研究从表型层面向分子机制层面推进。该文梳理了国内外学者近年的研究以及笔者所在课题组的研究成果,提出牙颌面骨畸形机制研究“一体两翼”模式,即牙颌面骨畸形为“一体”,颌面骨发育与畸形致病机制为“一翼”,牙-牙周复合体稳态与应激改建机制为“另一翼”;该模式的提出旨在系统性研究疾病的发生发展,探索临床干预新思路。近年的相关研究运用前沿技术从“两翼”出发探究“一体”的机制:一方面,牙颌面骨的胚胎发育来源复杂,组成型条件性模式动物成为研究关键细胞中关键因子功能的重要新策略;另一方面,牙-牙周复合体的成体改建最为频繁,诱导型条件性模式动物为模型时程精准控制提供了技术支持。随着单细胞测序与谱系示踪技术的开发,组织特异性干细胞因其原位、特化的特征渐受青睐,越来越多的研究者开始关注其特征功能,这一发展趋势十分契合“一体两翼”的研究模式,有望加快牙颌面骨畸形的理论基础建设与应用转化。该文就牙颌面骨畸形机制“一体两翼”的研究模式进行述评。
中图分类号:
江凌勇. 牙颌面骨畸形机制研究的现状与发展[J]. 上海交通大学学报(医学版), 2024, 44(6): 663-675.
JIANG Lingyong. Status and advances in the mechanism research on dento-maxillofacial skeletal abnormalities[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(6): 663-675.
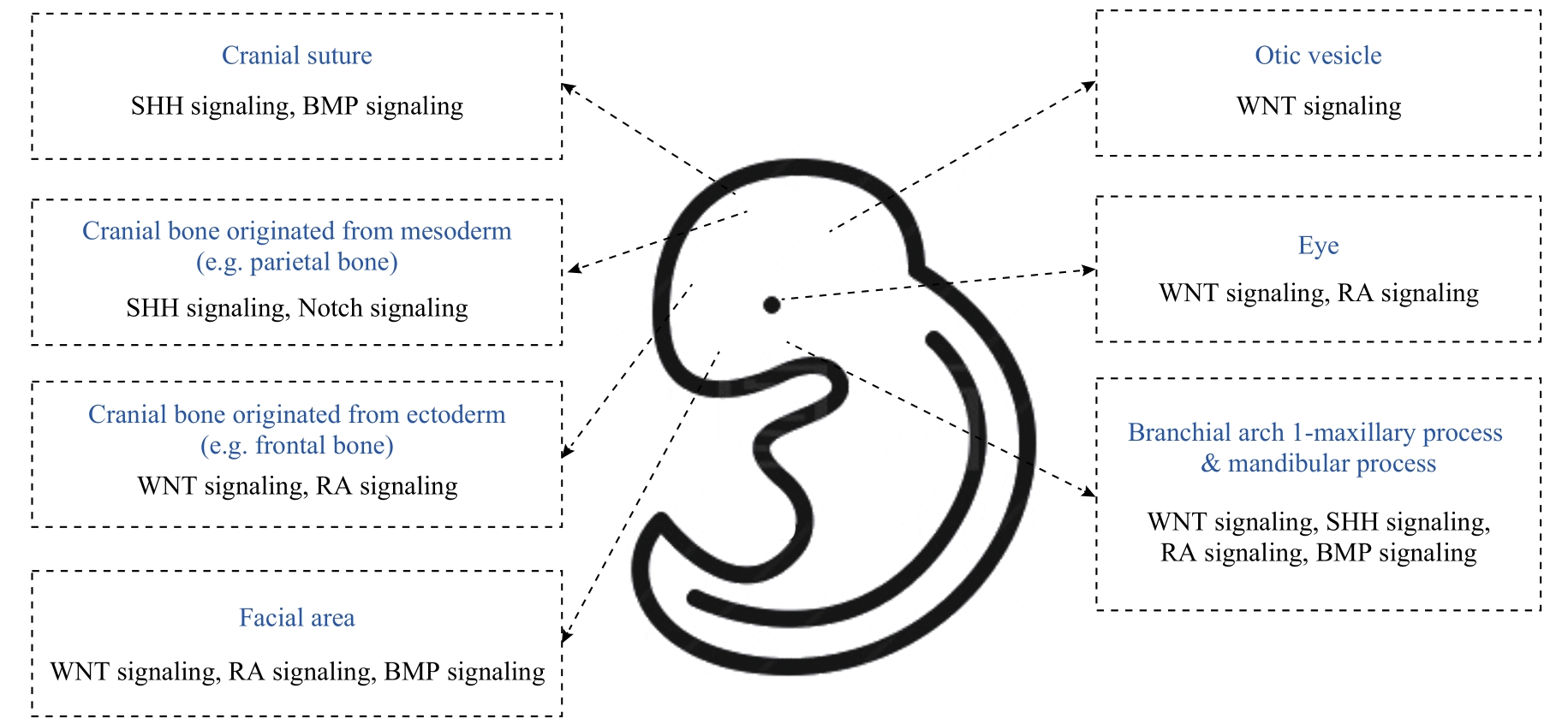
图2 LacZ 工具鼠实验提示WNT信号、SHH信号、RA信号、BMP信号、Notch信号参与牙颌面组织发育形成的示意图
Fig 2 Schematic diagram of the roles of WNT signaling, SHH signaling, RA signaling, BMP signaling, and Notch signaling during dento-maxillofacial development using LacZ tool mice
| Key cell lineage (marker-Cre) | Gene | Pathway | Characteristic of abnormalities | Related phenotype, syndrome or disease | Reference |
|---|---|---|---|---|---|
| NCC (Wnt1-Cre) | Med23 | WNT signaling | ③④ | Pierre Robin syndrome, micrognathia, cleft palate | [ |
| NCC (Wnt1-Cre) | Fgf18 | WNT signaling | ①③④ | Micrognathia, cleft palate, hypoplastic craniofacial bones | [ |
| NCC (Wnt1-Cre) | Six1 | WNT signaling, BMP signaling | ③④ | Branchio-oto-renal syndrome, micrognathia, cleft palate with ankyloglossia | [ |
| NCC (Wnt1-Cre) | Bmp2 | BMP signaling | ③④ | Pierre Robin syndrome | [ |
| NCC (Wnt1-Cre) | Bmp4 | BMP signaling | ②③④ | Severe deformation of molar buds, palate, and maxilla-mandibular bony structures; defected Meckel's cartilage | [ |
| NCC (Wnt1-Cre) | Foxf2 | SHH signaling | ④ | Cleft palate | [ |
| NCC (Wnt1-Cre) | Setdb1 | BMP signaling, WNT signaling | ④ | Cleft palate | [ |
| NCC (Wnt1-Cre) | Ift20 | WNT signaling | ①③④ | Death shortly after birth due to difficulties in feeding and breathing, severe craniofacial malformation, loss of craniofacial bones, frontonasal dysplasia, micrognathia, cleft palate | [ |
| NCC (Wnt1-Cre, Sox9-Cre) | G9a | SHH signaling | Wnt1: ①③; Sox9: ①② | Wnt1: death shortly after birth, shortened maxilla, restricted airway, frontonasal dysplasia Sox9: death shortly after birth, cranial skeletal dysplasia, smaller tooth germ, impaired tooth inner enamel epithelium | [ |
| NCC (Wnt1-Cre) | Tak1 | FGF signaling | ③④ | Pierre Robin syndrome, micrognathia, abnormal tongue, cleft palate | [ |
| NCC (Wnt1-Cre) | Tgfbr2 | FGF signaling | ‒ | Abnormal tongue | [ |
| NCC (Wnt1-Cre) | Nell1 | WNT signaling | ① | Craniosynostosis | [ |
| NCC (Wnt1-Cre) | Dlx3 | WNT signaling | ①③ | Tricho-dento-osseous syndrome, defected frontal bone and mandible | [ |
| NCC (Wnt1-Cre) | Yap/Taz | WNT signaling | ①③ | Neural tube malformation, craniofacial vascular malformation, mandibular abnormalities | [ |
| NCC (Wnt1-Cre) | Tfap2 | WNT signaling, RA signaling | ①③④ | Branchio-oto-renal syndrome, midface cleft, defected craniofacial bone | [ |
| NCC (Wnt1-Cre) | Twist1 | FGF signaling | ① | Defected craniofacial bone | [ |
| NCC (Wnt1-Cre) | Brca1/2 | P53 signaling | ①④ | Craniostenosis, cleft palate, defected craniofacial bone | [ |
| NCC (Wnt1-Cre) | Smo | SHH signaling | ① | Defected craniofacial bone | [ |
| NCC (Wnt1-Cre) | Ldb1 | WNT signaling | ④ | Cleft palate | [ |
| NCC (Wnt1-Cre) | Osx | FGF signaling | ①③ | Micrognathia, defected craniofacial bone | [ |
| First branchial arch mesenchymal cell (Pax2-Cre) | Bmp4 | BMP signaling | ‒ | Bilateral hyperplastic tissues | [ |
| First branchial arch mesenchymal cell (Hand2-Cre) | Twist1 | FGF signaling | ③④ | Micrognathia, cleft palate | [ |
| Mesenchymal cell (Twist2-Cre) | Fgf18 | WNT signaling | ①③ | Micrognathia, defected craniofacial bone | [ |
| Mesenchymal cell (Twist1-Cre) | Twist1 | FGF signaling | ② | Defected dentin and enamel, tooth abnormalities | [ |
| Osteoblast (Prx1-Cre) | Ift20 | WNT signaling | ① | Craniostenosis | [ |
| Osteoblast (Osx-Cre, Col1-Cre) | Fgfr3 | WNT signaling | ① | Osx: CATSHL syndrome, frontonasal dysplasia Col1: CATSHL syndrome | [ |
| Osteoblast (Osx-Cre) | Rar | RA signaling | ①③ | Vitamin A deficiency, micrognathia, frontonasal dysplasia | [ |
| Osteoblast (Prx1-Cre, Osx-Cre) | Ror2 | BMP signaling, STAT signaling | ①②③ | Robinow syndrome, brachyrhinia | [ |
| Osteoblast (Prx1-Cre, Osx-Cre) | Stat3 | STAT signaling | ①③ | AD-HIES syndrome, defected craniofacial bone | [ |
| Osteoblast/chondroblast (Dermo1-Cre, Col2a1-Cre, Prx1-Cre, Osx-Cre) | Cbfb | WNT signaling | ①②③ | Dermo1 & Col2a1: Cleidocranial dysostosis, hypomineralized craniofacial bones, clavicle dysplasia Prx1: Cleidocranial dysostosis, hypomineralized parietal bones, clavicle missing Osx: Cleidocranial dysostosis, hypomineralized parietal bones, tooth deformities | [ |
| Osteoblast/chondroblast (Prx1-Cre, Col2-Cre) | Recql4 | P53 signaling | ①② | Rothmund-Thomson syndrome, Baller-Gerold syndrome | [ |
| Chondroblast (Col2-Cre) | Yap/Taz | WNT signaling | ①④ | Defected craniofacial bone, cleft palate | [ |
| Osteoblast (Ocn-Cre) | Wls | WNT signaling | ①③ | Defected craniofacial bone, molar deformity | [ |
| Osteoblast (Ocn-Cre) | Ift20 | WNT signaling | ① | Osteopenia-like phenotypes in skulls | [ |
| Osteoblast (Osx-Cre) | Notch2 | Notch signaling | ①②③ | Hajdu-Cheney syndrome | [ |
| Osteoblast (Osx-Cre) | Fgfr2 | FGF signaling | ①②③ | Crouzon syndrome | [ |
| Osteoblast (Prrx1-Cre) | Gα | RANKL signaling | ① | Fibrous dysplasia | [ |
| Osteoblast (Osx-Cre) | Efnb1 | Unclear | ①③ | Larger cranial height, larger interorbital and nasal widths, smaller maxillary width | [ |
| Osteoblast (Osx-Cre) | Atg5 | MMP signaling | ① | Defected craniofacial bone | [ |
| Osteoblast (Osx-Cre) | Fip200 | MMP signaling | ① | Defected craniofacial bone | [ |
| Epithelial cell (Krt14-Cre、Eiia-Cre) | Wnt10a | WNT signaling | ② | Taurodontism | [ |
| Epithelial cell (Krt14-Cre) | Tgfbr2 | WNT signaling | ④ | Soft palate cleft | [ |
| Epithelial cell (Krt14-Cre) | Dlx3 | WNT signaling | ② | Hypomineralized enamel | [ |
| Epithelial cell (Krt14-Cre) | Fgfr2 | FGF signaling | ② | Retarded tooth formation, cleft palate | [ |
| Epithelial cell (Shh-Cre) | Wls | WNT signaling | ② | Defective ameloblast and odontoblast differentiation | [ |
| CNC-derived cell subset in the developing palatal mesenchyme (Osr2-Cre) | β-catenin | WNT signaling | ④ | Cleft palate | [ |
| CNC-derived cell subset in the developing palatal mesenchyme (Osr2-Cre) | Runx2 | RA signaling | ④ | Cleft palate | [ |
| Pharyngeal endoderm cell (Foxg1-Cre) | Tbx1 | Unclear | ①③ | Velo-cardio-facial syndrome | [ |
表1 条件性基因编辑小鼠鉴定得到的牙颌面关键细胞谱系及其相关畸形致病基因
Tab 1 Key cell lineages and related genes responsible for dento-maxillofacial abnormalities through conditional gene-edited mice
| Key cell lineage (marker-Cre) | Gene | Pathway | Characteristic of abnormalities | Related phenotype, syndrome or disease | Reference |
|---|---|---|---|---|---|
| NCC (Wnt1-Cre) | Med23 | WNT signaling | ③④ | Pierre Robin syndrome, micrognathia, cleft palate | [ |
| NCC (Wnt1-Cre) | Fgf18 | WNT signaling | ①③④ | Micrognathia, cleft palate, hypoplastic craniofacial bones | [ |
| NCC (Wnt1-Cre) | Six1 | WNT signaling, BMP signaling | ③④ | Branchio-oto-renal syndrome, micrognathia, cleft palate with ankyloglossia | [ |
| NCC (Wnt1-Cre) | Bmp2 | BMP signaling | ③④ | Pierre Robin syndrome | [ |
| NCC (Wnt1-Cre) | Bmp4 | BMP signaling | ②③④ | Severe deformation of molar buds, palate, and maxilla-mandibular bony structures; defected Meckel's cartilage | [ |
| NCC (Wnt1-Cre) | Foxf2 | SHH signaling | ④ | Cleft palate | [ |
| NCC (Wnt1-Cre) | Setdb1 | BMP signaling, WNT signaling | ④ | Cleft palate | [ |
| NCC (Wnt1-Cre) | Ift20 | WNT signaling | ①③④ | Death shortly after birth due to difficulties in feeding and breathing, severe craniofacial malformation, loss of craniofacial bones, frontonasal dysplasia, micrognathia, cleft palate | [ |
| NCC (Wnt1-Cre, Sox9-Cre) | G9a | SHH signaling | Wnt1: ①③; Sox9: ①② | Wnt1: death shortly after birth, shortened maxilla, restricted airway, frontonasal dysplasia Sox9: death shortly after birth, cranial skeletal dysplasia, smaller tooth germ, impaired tooth inner enamel epithelium | [ |
| NCC (Wnt1-Cre) | Tak1 | FGF signaling | ③④ | Pierre Robin syndrome, micrognathia, abnormal tongue, cleft palate | [ |
| NCC (Wnt1-Cre) | Tgfbr2 | FGF signaling | ‒ | Abnormal tongue | [ |
| NCC (Wnt1-Cre) | Nell1 | WNT signaling | ① | Craniosynostosis | [ |
| NCC (Wnt1-Cre) | Dlx3 | WNT signaling | ①③ | Tricho-dento-osseous syndrome, defected frontal bone and mandible | [ |
| NCC (Wnt1-Cre) | Yap/Taz | WNT signaling | ①③ | Neural tube malformation, craniofacial vascular malformation, mandibular abnormalities | [ |
| NCC (Wnt1-Cre) | Tfap2 | WNT signaling, RA signaling | ①③④ | Branchio-oto-renal syndrome, midface cleft, defected craniofacial bone | [ |
| NCC (Wnt1-Cre) | Twist1 | FGF signaling | ① | Defected craniofacial bone | [ |
| NCC (Wnt1-Cre) | Brca1/2 | P53 signaling | ①④ | Craniostenosis, cleft palate, defected craniofacial bone | [ |
| NCC (Wnt1-Cre) | Smo | SHH signaling | ① | Defected craniofacial bone | [ |
| NCC (Wnt1-Cre) | Ldb1 | WNT signaling | ④ | Cleft palate | [ |
| NCC (Wnt1-Cre) | Osx | FGF signaling | ①③ | Micrognathia, defected craniofacial bone | [ |
| First branchial arch mesenchymal cell (Pax2-Cre) | Bmp4 | BMP signaling | ‒ | Bilateral hyperplastic tissues | [ |
| First branchial arch mesenchymal cell (Hand2-Cre) | Twist1 | FGF signaling | ③④ | Micrognathia, cleft palate | [ |
| Mesenchymal cell (Twist2-Cre) | Fgf18 | WNT signaling | ①③ | Micrognathia, defected craniofacial bone | [ |
| Mesenchymal cell (Twist1-Cre) | Twist1 | FGF signaling | ② | Defected dentin and enamel, tooth abnormalities | [ |
| Osteoblast (Prx1-Cre) | Ift20 | WNT signaling | ① | Craniostenosis | [ |
| Osteoblast (Osx-Cre, Col1-Cre) | Fgfr3 | WNT signaling | ① | Osx: CATSHL syndrome, frontonasal dysplasia Col1: CATSHL syndrome | [ |
| Osteoblast (Osx-Cre) | Rar | RA signaling | ①③ | Vitamin A deficiency, micrognathia, frontonasal dysplasia | [ |
| Osteoblast (Prx1-Cre, Osx-Cre) | Ror2 | BMP signaling, STAT signaling | ①②③ | Robinow syndrome, brachyrhinia | [ |
| Osteoblast (Prx1-Cre, Osx-Cre) | Stat3 | STAT signaling | ①③ | AD-HIES syndrome, defected craniofacial bone | [ |
| Osteoblast/chondroblast (Dermo1-Cre, Col2a1-Cre, Prx1-Cre, Osx-Cre) | Cbfb | WNT signaling | ①②③ | Dermo1 & Col2a1: Cleidocranial dysostosis, hypomineralized craniofacial bones, clavicle dysplasia Prx1: Cleidocranial dysostosis, hypomineralized parietal bones, clavicle missing Osx: Cleidocranial dysostosis, hypomineralized parietal bones, tooth deformities | [ |
| Osteoblast/chondroblast (Prx1-Cre, Col2-Cre) | Recql4 | P53 signaling | ①② | Rothmund-Thomson syndrome, Baller-Gerold syndrome | [ |
| Chondroblast (Col2-Cre) | Yap/Taz | WNT signaling | ①④ | Defected craniofacial bone, cleft palate | [ |
| Osteoblast (Ocn-Cre) | Wls | WNT signaling | ①③ | Defected craniofacial bone, molar deformity | [ |
| Osteoblast (Ocn-Cre) | Ift20 | WNT signaling | ① | Osteopenia-like phenotypes in skulls | [ |
| Osteoblast (Osx-Cre) | Notch2 | Notch signaling | ①②③ | Hajdu-Cheney syndrome | [ |
| Osteoblast (Osx-Cre) | Fgfr2 | FGF signaling | ①②③ | Crouzon syndrome | [ |
| Osteoblast (Prrx1-Cre) | Gα | RANKL signaling | ① | Fibrous dysplasia | [ |
| Osteoblast (Osx-Cre) | Efnb1 | Unclear | ①③ | Larger cranial height, larger interorbital and nasal widths, smaller maxillary width | [ |
| Osteoblast (Osx-Cre) | Atg5 | MMP signaling | ① | Defected craniofacial bone | [ |
| Osteoblast (Osx-Cre) | Fip200 | MMP signaling | ① | Defected craniofacial bone | [ |
| Epithelial cell (Krt14-Cre、Eiia-Cre) | Wnt10a | WNT signaling | ② | Taurodontism | [ |
| Epithelial cell (Krt14-Cre) | Tgfbr2 | WNT signaling | ④ | Soft palate cleft | [ |
| Epithelial cell (Krt14-Cre) | Dlx3 | WNT signaling | ② | Hypomineralized enamel | [ |
| Epithelial cell (Krt14-Cre) | Fgfr2 | FGF signaling | ② | Retarded tooth formation, cleft palate | [ |
| Epithelial cell (Shh-Cre) | Wls | WNT signaling | ② | Defective ameloblast and odontoblast differentiation | [ |
| CNC-derived cell subset in the developing palatal mesenchyme (Osr2-Cre) | β-catenin | WNT signaling | ④ | Cleft palate | [ |
| CNC-derived cell subset in the developing palatal mesenchyme (Osr2-Cre) | Runx2 | RA signaling | ④ | Cleft palate | [ |
| Pharyngeal endoderm cell (Foxg1-Cre) | Tbx1 | Unclear | ①③ | Velo-cardio-facial syndrome | [ |
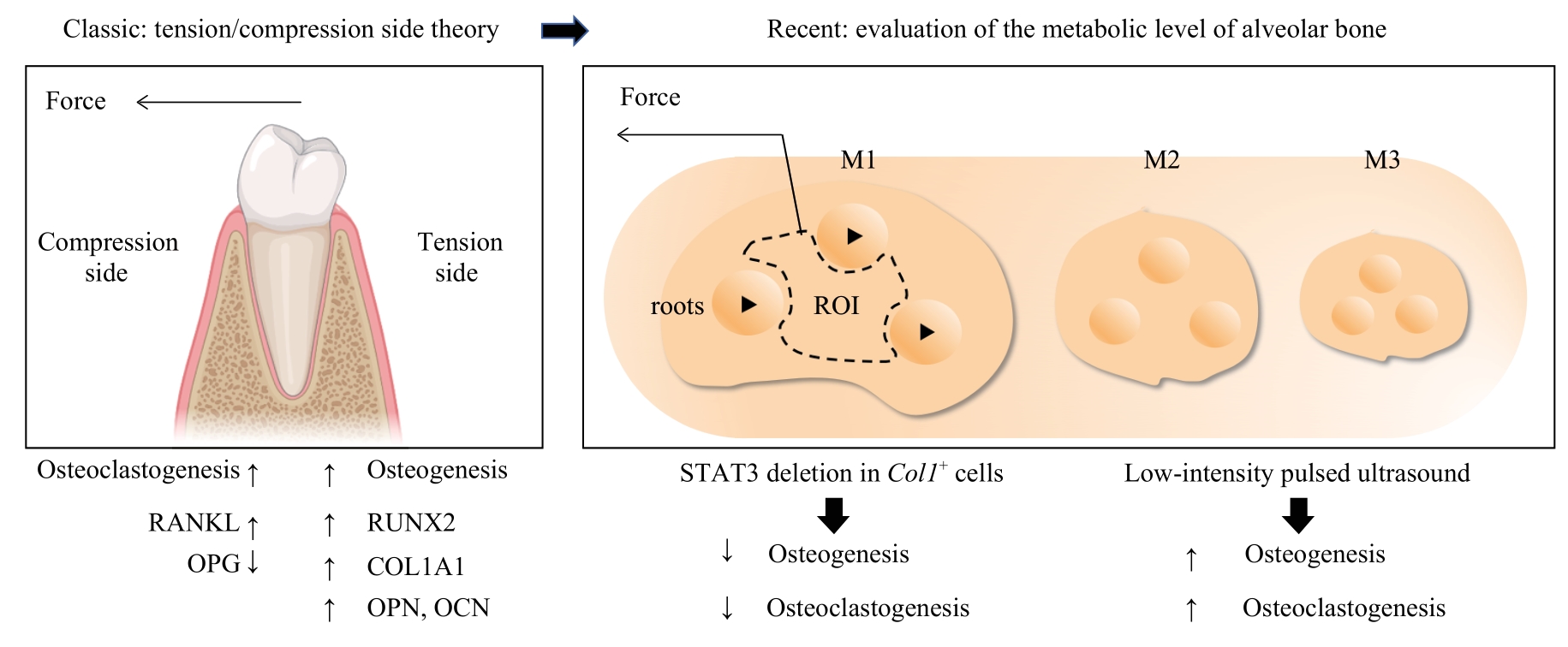
图3 应力下牙-牙周复合体整体代谢水平评价模式的转变Note: OPG—osteoprotegerin; M1/M2/M3—molar 1/2/3; ROI—region of interest.
Fig 3 Transform of evaluation of the general metabolic level of tooth-periodontal complex under stress
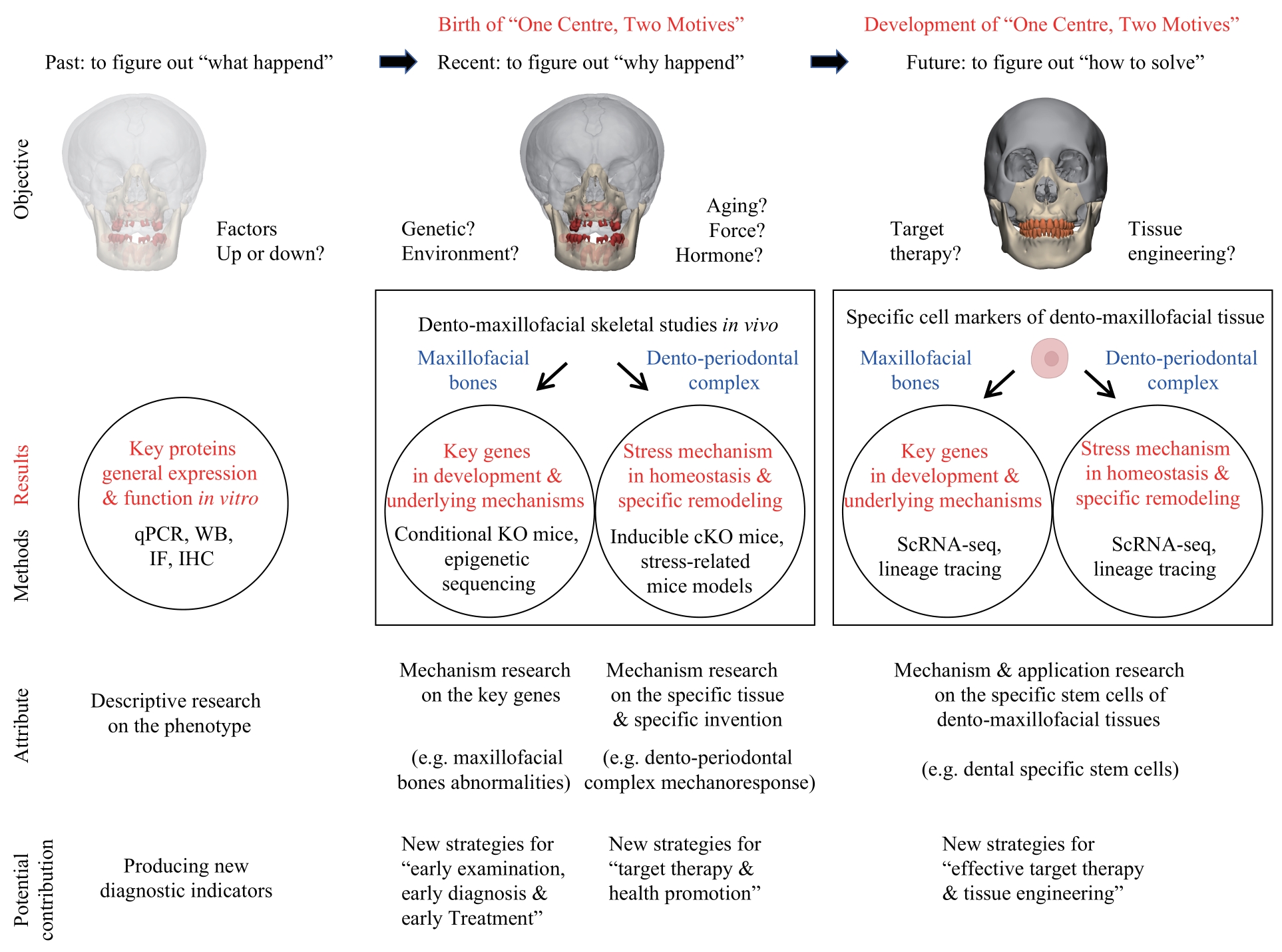
图5 牙颌面骨畸形“一体两翼”研究模式的现状、发展与展望Note:qPCR—quantitative polymerase chain reaction; WB—Western blotting; IF—immunofluorescence; IHC—immunohistochemistry; KO—knockout; cKO—conditional knockout; ScRNA-seq—single cell RNA sequencing.
Fig 5 Current status and future of the "One Centre, Two Motives" research mode for dento-maxillofacial skeletal abnormalities
| 1 | HEGGIE A A. Craniofacial disorders[J]. Aust Dent J, 2018, 63(Suppl 1): S58-S68. |
| 2 | PRUZINSKY T. Social and psychological effects of major craniofacial deformity[J]. Cleft Palate Craniofacial J, 1992, 29(6): 578-584. |
| 3 | ZHOU S R, DAI Q G, HUANG X R, et al. STAT3 is critical for skeletal development and bone homeostasis by regulating osteogenesis[J]. Nat Commun, 2021, 12(1): 6891. |
| 4 | GONG X, SUN S, YANG Y, et al. Osteoblastic STAT3 is crucial for orthodontic force driving alveolar bone remodeling and tooth movement[J]. J Bone Miner Res, 2023, 38(1): 214-227. |
| 5 | JIN A, XU H, GAO X, et al. ScRNA-seq reveals a distinct osteogenic progenitor of alveolar bone[J]. J Dent Res, 2023, 102(6): 645-655. |
| 6 | MANZANARES M, NIETO M A. A celebration of the new head and an evaluation of the new mouth[J]. Neuron, 2003, 37(6): 895-898. |
| 7 | HELMS J A, SCHNEIDER R A. Cranial skeletal biology[J]. Nature, 2003, 423(6937): 326-331. |
| 8 | WILKIE A O, MORRISS-KAY G M. Genetics of craniofacial development and malformation[J]. Nat Rev Genet, 2001, 2(6): 458-468. |
| 9 | CUGURRA A, MAMULADZE T, RUSTENHOVEN J, et al. Skull and vertebral bone marrow are myeloid cell reservoirs for the meninges and CNS parenchyma[J]. Science, 2021, 373(6553): eabf7844. |
| 10 | BOK S, YALLOWITZ A R, SUN J, et al. A multi-stem cell basis for craniosynostosis and calvarial mineralization[J]. Nature, 2023, 621(7980): 804-812. |
| 11 | YU M F, MA L, YUAN Y, et al. Cranial suture regeneration mitigates skull and neurocognitive defects in craniosynostosis[J]. Cell, 2021, 184(1): 243-256.e18. |
| 12 | YEWENG S J, HUANG S F, REN L J. Orthodontics in China[J]. J Orthod, 2002, 29(1): 62-65. |
| 13 | PATIL S, RAO R S, MAJUMDAR B. Single gene disorders with craniofacial and oral manifestations[J]. J Contemp Dent Pract, 2014, 15(5): 659-671. |
| 14 | PATEL N C, GALLAGHER J L, TORGERSON T R, et al. Successful haploidentical donor hematopoietic stem cell transplant and restoration of STAT3 function in an adolescent with autosomal dominant hyper-IgE syndrome[J]. J Clin Immunol, 2015, 35(5): 479-485. |
| 15 | MANI P, JARRELL A, MYERS J, et al. Visualizing canonical Wnt signaling during mouse craniofacial development[J]. Dev Dyn, 2010, 239(1): 354-363. |
| 16 | LIU B, HUNTER D J, ROOKER S, et al. Wnt signaling promotes Müller cell proliferation and survival after injury[J]. Invest Ophthalmol Vis Sci, 2013, 54(1): 444-453. |
| 17 | LIM W H, LIU B, CHENG D, et al. Wnt signaling regulates homeostasis of the periodontal ligament[J]. J Periodontal Res, 2014, 49(6): 751-759. |
| 18 | DAO D Y, YANG X, FLICK L M, et al. Axin2 regulates chondrocyte maturation and axial skeletal development[J]. J Orthop Res, 2010, 28(1): 89-95. |
| 19 | LOHI M, TUCKER A S, SHARPE P T. Expression of Axin2 indicates a role for canonical Wnt signaling in development of the crown and root during pre- and postnatal tooth development[J]. Dev Dyn, 2010, 239(1): 160-167. |
| 20 | MENG Q H, MONGAN M, WANG J J, et al. Repression of MAP3K1 expression and JNK activity by canonical Wnt signaling[J]. Dev Biol, 2018, 440(2): 129-136. |
| 21 | NAKASHIMA M, TANESE N, ITO M, et al. A novel gene, GliH1, with homology to the Gli zinc finger domain not required for mouse development[J]. Mech Dev, 2002, 119(1): 21-34. |
| 22 | GUSTAFSSON M K, PAN H, PINNEY D F, et al. Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification[J]. Genes Dev, 2002, 16(1): 114-126. |
| 23 | PATAPOUTIAN A, MINER J H, LYONS G E, et al. Isolated sequences from the linked Myf-5 and MRF4 genes drive distinct patterns of muscle-specific expression in transgenic mice[J]. Development, 1993, 118(1): 61-69. |
| 24 | LOHNES D, MARK M, MENDELSOHN C, et al. Function of the retinoic acid receptors (RARs) during development: (Ⅰ) Craniofacial and skeletal abnormalities in RAR double mutants[J]. Development, 1994, 120(10): 2723-2748. |
| 25 | YU X, KAWAKAMI H, TAHARA N, et al. Expression of Noggin and Gremlin1 and its implications in fine-tuning BMP activities in mouse cartilage tissues[J]. J Orthop Res, 2017, 35(8): 1671-1682. |
| 26 | NACKE S, SCHÄFER R, HABRÉ DE ANGELIS M, et al. Mouse mutant “rib-vertebrae” (rv): a defect in somite polarity[J]. Dev Dyn, 2000, 219(2): 192-200. |
| 27 | LI Q W, XU R S, LEI K X, et al. Insights into skeletal stem cells[J]. Bone Res, 2022, 10(1): 61. |
| 28 | BAEK W Y, KIM Y J, CROMBRUGGHE B D, et al. Osterix is required for cranial neural crest-derived craniofacial bone formation[J]. Biochem Biophys Res Commun, 2013, 432(1): 188-192. |
| 29 | XIAO M, ZHANG W J, LIU W, et al. Osteocytes regulate neutrophil development through IL-19: a potent cytokine for neutropenia treatment[J]. Blood, 2021, 137(25): 3533-3547. |
| 30 | QIN X, JIANG Q, MATSUO Y, et al. Cbfb regulates bone development by stabilizing Runx family proteins[J]. J Bone Miner Res, 2015, 30(4): 706-714. |
| 31 | YANG Y L, CHUNG M R, ZHOU S R, et al. STAT3 controls osteoclast differentiation and bone homeostasis by regulating NFATc1 transcription[J]. J Biol Chem, 2019, 294(42): 15395-15407. |
| 32 | XU J, CHEN M L, YAN Y N, et al. The effects of altered BMP4 signaling in first branchial-arch-derived murine embryonic orofacial tissues[J]. Int J Oral Sci, 2021, 13(1): 40. |
| 33 | HAN X, FENG J F, GUO T W, et al. Runx2-Twist1 interaction coordinates cranial neural crest guidance of soft palate myogenesis[J]. Elife, 2021, 10: e62387. |
| 34 | CHEN J Q, SHI Y, REGAN J, et al. Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice[J]. PLoS One, 2014, 9(1): e85161. |
| 35 | DASH S, BHATT S, FALCON K T, et al. Med23 regulates Sox9 expression during craniofacial development[J]. J Dent Res, 2021, 100(4): 406-414. |
| 36 | YUE M H, LAN Y, LIU H, et al. Tissue-specific analysis of Fgf18 gene function in palate development[J]. Dev Dyn, 2021, 250(4): 562-573. |
| 37 | LUO S Y, LIU Z X, BIAN Q, et al. Ectomesenchymal Six1 controls mandibular skeleton formation[J]. Front Genet, 2023, 14: 1082911. |
| 38 | CHEN Y X, WANG Z S, CHEN Y P, et al. Conditional deletion of Bmp2 in cranial neural crest cells recapitulates Pierre Robin sequence in mice[J]. Cell Tissue Res, 2019, 376(2): 199-210. |
| 39 | XU J Y, LIU H, LAN Y, et al. A Shh-Foxf-Fgf18-Shh molecular circuit regulating palate development[J]. PLoS Genet, 2016, 12(1): e1005769. |
| 40 | KANO S, HIGASHIHORI N, THIHA P, et al. The role of the histone methyltransferase SET domain bifurcated 1 during palatal development[J]. Biochem Biophys Res Commun, 2022, 598: 74-80. |
| 41 | NODA K, KITAMI M, KITAMI K, et al. Canonical and noncanonical intraflagellar transport regulates craniofacial skeletal development[J]. Proc Natl Acad Sci U S A, 2016, 113(19): E2589-E2597. |
| 42 | YAMAGUCHI H, TERAJIMA M, KITAMI M, et al. IFT20 is critical for collagen biosynthesis in craniofacial bone formation[J]. Biochem Biophys Res Commun, 2020, 533(4): 739-744. |
| 43 | HIGASHIHORI N, LEHNERTZ B, SAMPAIO A, et al. Methyltransferase G9A regulates osteogenesis via Twist gene repression[J]. J Dent Res, 2017, 96(10): 1136-1144. |
| 44 | IDENO H, NAKASHIMA K, KOMATSU K, et al. G9a is involved in the regulation of cranial bone formation through activation of Runx2 function during development[J]. Bone, 2020, 137: 115332. |
| 45 | KAMIUNTEN T, IDENO H, SHIMADA A, et al. Essential roles of G9a in cell proliferation and differentiation during tooth development[J]. Exp Cell Res, 2017, 357(2): 202-210. |
| 46 | SONG Z C, LIU C, IWATA J, et al. Mice with Tak1 deficiency in neural crest lineage exhibit cleft palate associated with abnormal tongue development[J]. J Biol Chem, 2013, 288(15): 10440-10450. |
| 47 | HOSOKAWA R, OKA K, YAMAZA T, et al. TGF-β mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis[J]. Dev Biol, 2010, 341(1): 186-195. |
| 48 | CHEN X Y, WANG H M, YU M L, et al. Cumulative inactivation of Nell-1 in Wnt1 expressing cell lineages results in craniofacial skeletal hypoplasia and postnatal hydrocephalus[J]. Cell Death Differ, 2020, 27(4): 1415-1430. |
| 49 | DUVERGER O, OHARA T, BIBLE P W, et al. DLX3-dependent regulation of ion transporters and carbonic anhydrases is crucial for enamel mineralization[J]. J Bone Miner Res, 2017, 32(3): 641-653. |
| 50 | WANG J, XIAO Y, HSU C W, et al. Yap and Taz play a crucial role in neural crest-derived craniofacial development[J]. Development, 2016, 143(3): 504-515. |
| 51 | NGUYEN T T, MITCHELL J M, KIEL M D, et al. TFAP2 paralogs regulate midfacial development in part through a conserved ALX genetic pathway[J]. Development, 2024, 151(1): dev202095. |
| 52 | BILDSOE H, LOEBEL D A, JONES V J, et al. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo[J]. Dev Biol, 2009, 331(2): 176-188. |
| 53 | KITAMI K, KITAMI M, KAKU M, et al. BRCA1 and BRCA2 tumor suppressors in neural crest cells are essential for craniofacial bone development[J]. PLoS Genet, 2018, 14(5): e1007340. |
| 54 | JEONG J, MAO J H, TENZEN T, et al. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia[J]. Genes Dev, 2004, 18(8): 937-951. |
| 55 | ALMAIDHAN A, CESARIO J, LANDIN MALT A, et al. Neural crest-specific deletion of Ldb1 leads to cleft secondary palate with impaired palatal shelf elevation[J]. BMC Dev Biol, 2014, 14: 3. |
| 56 | ZHANG Y P, BLACKWELL E L, MCKNIGHT M T, et al. Specific inactivation of Twist1 in the mandibular arch neural crest cells affects the development of the ramus and reveals interactions with Hand2[J]. Dev Dyn, 2012, 241(5): 924-940. |
| 57 | MENG T, HUANG Y Y, WANG S Z, et al. Twist1 is essential for tooth morphogenesis and odontoblast differentiation[J]. J Biol Chem, 2015, 290(49): 29593-29602. |
| 58 | YAMAGUCHI H, KITAMI M, UCHIMA KOECKLIN K H, et al. Temporospatial regulation of intraflagellar transport is required for the endochondral ossification in mice[J]. Dev Biol, 2022, 482: 91-100. |
| 59 | BIOSSE DUPLAN M, DAMBROISE E, ESTIBALS V, et al. An Fgfr3-activating mutation in immature murine osteoblasts affects the appendicular and craniofacial skeleton[J]. Dis Model Mech, 2021, 14(4): dmm048272. |
| 60 | SUN X D, ZHANG R B, CHEN H G, et al. Fgfr3 mutation disrupts chondrogenesis and bone ossification in zebrafish model mimicking CATSHL syndrome partially via enhanced Wnt/β-catenin signaling[J]. Theranostics, 2020, 10(16): 7111-7130. |
| 61 | DAI Q, SUN S, JIN A, et al. Osteoblastic RAR inhibition causes VAD-like craniofacial skeletal deformity[J]. J Dent Res, 2023, 102(6): 667-677. |
| 62 | LEI L Z, HUANG Z W, FENG J Y, et al. Loss of receptor tyrosine kinase-like orphan receptor 2 impairs the osteogenesis of mBMSCs by inhibiting signal transducer and activator of transcription 3[J]. Stem Cell Res Ther, 2020, 11(1): 137. |
| 63 | YADAV P S, FENG S H, CONG Q, et al. Stat3 loss in mesenchymal progenitors causes Job syndrome-like skeletal defects by reducing Wnt/β-catenin signaling[J]. Proc Natl Acad Sci U S A, 2021, 118(26): e2020100118. |
| 64 | CHEN W, MA J Q, ZHU G C, et al. Cbfβ deletion in mice recapitulates cleidocranial dysplasia and reveals multiple functions of Cbfβ required for skeletal development[J]. Proc Natl Acad Sci U S A, 2014, 111(23): 8482-8487. |
| 65 | TIAN F, WU M R, DENG L F, et al. Core binding factor beta (Cbfβ) controls the balance of chondrocyte proliferation and differentiation by upregulating Indian hedgehog (Ihh) expression and inhibiting parathyroid hormone-related protein receptor (PPR) expression in postnatal cartilage and bone formation[J]. J Bone Miner Res, 2014, 29(7): 1564-1574. |
| 66 | LU L C, HARUTYUNYAN K, JIN W D, et al. RECQL4 regulates p53 function in vivo during skeletogenesis[J]. J Bone Miner Res, 2015, 30(6): 1077-1089. |
| 67 | GOODWIN A F, CHEN C P, VO N T, et al. YAP/TAZ regulate elevation and bone formation of the mouse secondary palate[J]. J Dent Res, 2020, 99(12): 1387-1396. |
| 68 | LIM W H, LIU B, CHENG D, et al. Wnt signaling regulates pulp volume and dentin thickness[J]. J Bone Miner Res, 2014, 29(4): 892-901. |
| 69 | CANALIS E, GROSSMAN T R, CARRER M, et al. Antisense oligonucleotides targeting Notch2 ameliorate the osteopenic phenotype in a mouse model of Hajdu-Cheney syndrome[J]. J Biol Chem, 2020, 295(12): 3952-3964. |
| 70 | ZANOTTI S, YU J, SANJAY A, et al. Sustained Notch2 signaling in osteoblasts, but not in osteoclasts, is linked to osteopenia in a mouse model of Hajdu-Cheney syndrome[J]. J Biol Chem, 2017, 292(29): 12232-12244. |
| 71 | LEE K K L, PESKETT E, QUINN C M, et al. Overexpression of Fgfr2c causes craniofacial bone hypoplasia and ameliorates craniosynostosis in the Crouzon mouse[J]. Dis Model Mech, 2018, 11(11): dmm035311. |
| 72 | KARUPPAIAH K, YU K, LIM J, et al. FGF signaling in the osteoprogenitor lineage non-autonomously regulates postnatal chondrocyte proliferation and skeletal growth[J]. Development, 2016, 143(10): 1811-1822. |
| 73 | ZHAO X F, DENG P, IGLESIAS-BARTOLOME R, et al. Expression of an active Gαs mutant in skeletal stem cells is sufficient and necessary for fibrous dysplasia initiation and maintenance[J]. Proc Natl Acad Sci U S A, 2018, 115(3): E428-E437. |
| 74 | BEREZA S, YONG R, GRONTHOS S, et al. Craniomaxillofacial morphology in a murine model of ephrinB1 conditional deletion in osteoprogenitor cells[J]. Arch Oral Biol, 2022, 137: 105389. |
| 75 | THOMAS N, CHOI H K, WEI X X, et al. Autophagy regulates craniofacial bone acquisition[J]. Calcif Tissue Int, 2019, 105(5): 518-530. |
| 76 | XIONG Y, FANG Y, QIAN Y, et al. Wnt production in dental epithelium is crucial for tooth differentiation[J]. J Dent Res, 2019, 98(5): 580-588. |
| 77 | IWATA J I, SUZUKI A, YOKOTA T, et al. TGFβ regulates epithelial-mesenchymal interactions through WNT signaling activity to control muscle development in the soft palate[J]. Development, 2014, 141(4): 909-917. |
| 78 | DUVERGER O, ISAAC J, ZAH A, et al. In vivo impact of Dlx3 conditional inactivation in neural crest-derived craniofacial bones[J]. J Cell Physiol, 2013, 228(3): 654-664. |
| 79 | HOSOKAWA R, DENG X M, TAKAMORI K, et al. Epithelial-specific requirement of FGFR2 signaling during tooth and palate development[J]. J Exp Zool B Mol Dev Evol, 2009, 312B(4): 343-350. |
| 80 | JANEČKOVÁ E, FENG J F, GUO T W, et al. Canonical Wnt signaling regulates soft palate development by mediating ciliary homeostasis[J]. Development, 2023, 150(5): dev201189. |
| 81 | ARNOLD J S, WERLING U, BRAUNSTEIN E M, et al. Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations[J]. Development, 2006, 133(5): 977-987. |
| 82 | KATSIMBRI P. The biology of normal bone remodelling[J]. Eur J Cancer Care, 2017, 26(6). DOI: 10.1111/ecc.12740. |
| 83 | MALTHA J C, KUIJPERS-JAGTMAN A M. Mechanobiology of orthodontic tooth movement: an update[J]. J World Fed Orthod, 2023, 12(4): 156-160. |
| 84 | SAFFAR J L, LASFARGUES J J, CHERRUAU M. Alveolar bone and the alveolar process: the socket that is never stable[J]. Periodontol 2000, 1997, 13: 76-90. |
| 85 | RANSOM R C, CARTER A C, SALHOTRA A, et al. Mechanoresponsive stem cells acquire neural crest fate in jaw regeneration[J]. Nature, 2018, 563(7732): 514-521. |
| 86 | TANNE K, NAGATAKI T, MATSUBARA S, et al. Association between mechanical stress and bone remodeling[J]. J Osaka Univ Dent Sch, 1990, 30: 64-71. |
| 87 | SIBONGA J D. Spaceflight-induced bone loss: is there an osteoporosis risk?[J]. Curr Osteoporos Rep, 2013, 11(2): 92-98. |
| 88 | JIN A, HONG Y, YANG Y, et al. FOXO3 mediates tooth movement by regulating force-induced osteogenesis[J]. J Dent Res, 2022, 101(2): 196-205. |
| 89 | XU X, LIU S Y, LIU H, et al. Piezo channels: awesome mechanosensitive structures in cellular mechanotransduction and their role in bone[J]. Int J Mol Sci, 2021, 22(12): 6429. |
| 90 | WANG X Y, HUANG X R, GAO X, et al. Differentiation potential of periodontal Col1+ cells under orthodontic force[J]. Mechanobiol Med, 2024, 2(1): 100026. |
| 91 | WANG Y J, LI J, ZHOU J P, et al. Low-intensity pulsed ultrasound enhances bone marrow-derived stem cells-based periodontal regenerative therapies[J]. Ultrasonics, 2022, 121: 106678. |
| 92 | ZHOU J, ZHU Y L, AI D Q, et al. Low-intensity pulsed ultrasound regulates osteoblast-osteoclast crosstalk via EphrinB2/EphB4 signaling for orthodontic alveolar bone remodeling[J]. Front Bioeng Biotechnol, 2023, 11: 1192720. |
| 93 | YANG C Y, JEON H H, ALSHABAB A, et al. RANKL deletion in periodontal ligament and bone lining cells blocks orthodontic tooth movement[J]. Int J Oral Sci, 2018, 10(1): 3. |
| 94 | GRONTHOS S, MANKANI M, BRAHIM J, et al. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo[J]. Proc Natl Acad Sci U S A, 2000, 97(25): 13625-13630. |
| 95 | MIURA M, GRONTHOS S, ZHAO M R, et al. SHED: stem cells from human exfoliated deciduous teeth[J]. Proc Natl Acad Sci U S A, 2003, 100(10): 5807-5812. |
| 96 | WEI F L, SONG T L, DING G, et al. Functional tooth restoration by allogeneic mesenchymal stem cell-based bio-root regeneration in swine[J]. Stem Cells Dev, 2013, 22(12): 1752-1762. |
| 97 | LIU Y, YANG R L, LIU X B, et al. Hydrogen sulfide maintains mesenchymal stem cell function and bone homeostasis via regulation of Ca2+ channel sulfhydration[J]. Cell Stem Cell, 2014, 15(1): 66-78. |
| 98 | XUAN K, LI B, GUO H, et al. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth[J]. Sci Transl Med, 2018, 10(455): eaaf3227. |
| 99 | MARUYAMA T, JEONG J, SHEU T J, et al. Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration[J]. Nat Commun, 2016, 7: 10526. |
| 100 | WILK K, YEH S C A, MORTENSEN L J, et al. Postnatal calvarial skeletal stem cells expressing PRX1 reside exclusively in the calvarial sutures and are required for bone regeneration[J]. Stem Cell Reports, 2017, 8(4): 933-946. |
| 101 | ZHAO H, FENG J F, HO T V, et al. The suture provides a niche for mesenchymal stem cells of craniofacial bones[J]. Nat Cell Biol, 2015, 17(4): 386-396. |
| 102 | ORTINAU L C, WANG H, LEI K, et al. Identification of functionally distinct Mx1+αSMA+ periosteal skeletal stem cells[J]. Cell Stem Cell, 2019, 25(6): 784-796.e5. |
| 103 | WENG Y T, WANG H C, WU D, et al. A novel lineage of osteoprogenitor cells with dual epithelial and mesenchymal properties govern maxillofacial bone homeostasis and regeneration after MSFL[J]. Cell Res, 2022, 32(9): 814-830. |
| 104 | ZHANG N, BARRELL W B, LIU K J. Identification of distinct subpopulations of Gli1-lineage cells in the mouse mandible[J]. J Anat, 2023, 243(1): 90-99. |
| 105 | DING Y, MO C, GENG J, et al. Identification of periosteal osteogenic progenitors in jawbone[J]. J Dent Res, 2022, 101(9): 1101-1109. |
| 106 | ROGULJIC H, MATTHEWS B G, YANG W, et al. In vivo identification of periodontal progenitor cells[J]. J Dent Res, 2013, 92(8): 709-715. |
| 107 | YUAN X, PEI X, ZHAO Y, et al. A Wnt-responsive PDL population effectuates extraction socket healing[J]. J Dent Res, 2018, 97(7): 803-809. |
| 108 | BASSIR S H, GARAKANI S, WILK K, et al. Prx1 expressing cells are required for periodontal regeneration of the mouse incisor[J]. Front Physiol, 2019, 10: 591. |
| 109 | ZHAO H, FENG J F, SEIDEL K, et al. Secretion of Shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor[J]. Cell Stem Cell, 2014, 14(2): 160-173. |
| 110 | TAKAHASHI A, NAGATA M, GUPTA A, et al. Autocrine regulation of mesenchymal progenitor cell fates orchestrates tooth eruption[J]. Proc Natl Acad Sci U S A, 2019, 116(2): 575-580. |
| 111 | KAUKUA N, SHAHIDI M K, KONSTANTINIDOU C, et al. Glial origin of mesenchymal stem cells in a tooth model system[J]. Nature, 2014, 513(7519): 551-554. |
| 112 | CHEN H, FU H C, WU X, et al. Regeneration of pulpo-dentinal-like complex by a group of unique multipotent CD24a+ stem cells[J]. Sci Adv, 2020, 6(15): eaay1514. |
| 113 | ZHANG D, ZHANG S, WANG J, et al. LepR-expressing stem cells are essential for alveolar bone regeneration[J]. J Dent Res, 2020, 99(11): 1279-1286. |
| 114 | SHALEHIN N, SEKI Y, TAKEBE H, et al. Gli1+-PDL cells contribute to alveolar bone homeostasis and regeneration[J]. J Dent Res, 2022, 101(12): 1537-1543. |
| 115 | WU L D, LIU Z X, XIAO L, et al. The role of Gli1+ mesenchymal stem cells in osteogenesis of craniofacial bone[J]. Biomolecules, 2023, 13(9): 1351. |
| 116 | BUECHLER M B, PRADHAN R N, KRISHNAMURTY A T, et al. Cross-tissue organization of the fibroblast lineage[J]. Nature, 2021, 593(7860): 575-579. |
| 117 | LI W D, CAVELTI-WEDER C, ZHANG Y Y, et al. Long-term persistence and development of induced pancreatic beta cells generated by lineage conversion of acinar cells[J]. Nat Biotechnol, 2014, 32(12): 1223-1230. |
| 118 | TANG J, WANG H X, HUANG X Z, et al. Arterial Sca1+ vascular stem cells generate de novo smooth muscle for artery repair and regeneration[J]. Cell Stem Cell, 2020, 26(1): 81-96.e4. |
| 119 | SUN J, HU L L, BOK S, et al. A vertebral skeletal stem cell lineage driving metastasis[J]. Nature, 2023, 621(7979): 602-609. |
| 120 | JING J J, FENG J F, YUAN Y, et al. Spatiotemporal single-cell regulatory atlas reveals neural crest lineage diversification and cellular function during tooth morphogenesis[J]. Nat Commun, 2022, 13(1): 4803. |
| [1] | 周月, 程晨, 郑恩霖, 孟卓, 王鉴, 王青洁, 何勇宁, 孙锟. 利用Crispr/Cas9基因编辑系统在人胚胎干细胞中探索ELABELA的潜在新受体[J]. 上海交通大学学报(医学版), 2022, 42(9): 1258-1264. |
| [2] | 韩夏夏, 顾霜霜, 戴黛, 沈南. 应用CRISPR/Cas9介导的基因编辑系统研究B细胞中转录因子T-bet的调控作用[J]. 上海交通大学学报(医学版), 2022, 42(4): 433-442. |
| [3] | 赵艳娜, 邱荣, 沈南, 唐元家. 构建诱导型CRISPR/Cas9系统用于小鼠免疫细胞基因功能研究[J]. 上海交通大学学报(医学版), 2021, 41(3): 297-301. |
| [4] | 杨前昊,朱道宇,陈亦轩,高悠水,张长青. 哺乳动物雷帕霉素靶蛋白信号通路在骨稳态及相关疾病中作用的研究进展[J]. 上海交通大学学报(医学版), 2018, 38(11): 1391-. |
| [5] | 翁震 1,钮晓音 2,周俊松 1. CRISPR-Cas9技术在非肿瘤性血液病中的应用[J]. 上海交通大学学报(医学版), 2018, 38(11): 1396-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
