
上海交通大学学报(医学版) ›› 2021, Vol. 41 ›› Issue (4): 448-458.doi: 10.3969/j.issn.1674-8115.2021.04.006
收稿日期:2020-06-04
出版日期:2021-04-28
发布日期:2021-05-14
作者简介:顾琦晟 (1994—),男,硕士生;电子信箱: 基金资助:
Qi-sheng GU( ), Mi-li ZHANG, Can CAO, Ji-kun LI(
), Mi-li ZHANG, Can CAO, Ji-kun LI( )
)
Received:2020-06-04
Online:2021-04-28
Published:2021-05-14
Supported by:摘要:
目的·基于癌症基因组图谱数据库TCGA(The Cancer Genome Atlas)探讨胃癌可变剪接与肿瘤免疫的关系。方法·下载TCGA数据库中375例胃癌患者及32例配对癌旁正常组织的转录组、基因组数据和所有纳入研究的胃癌患者的临床信息,以及Spliceseq数据库452例胃癌及配对癌旁样本的剪接百分比数据。在GEO(Gene Expression Omnibus)数据库中下载包含192例胃癌患者转录组芯片数据的数据集GSE15459。利用ConcensusClusterPlus包对样本的可变剪接数据进行分型,通过竞争性基因集测试算法(Competitive Gene Set Test Accounting for Inter-gene Correlation,CAMERA)和基因集变异分析(Gene Set Variation Analysis, GSVA)对样本进行通路和基因集分析。采用CIBERSORT方法,通过反卷积分析胃癌样本的免疫微环境情况。结果·经过滤,纳入445例胃癌及配对癌旁组织正常样本进行分析,共涉及4 051个基因和8 649个剪接事件。胃癌组织与癌旁正常组织相比,有大量异常剪接事件发生。基于胃癌的不同可变剪接事件频率,将胃癌分为4个亚型。亚型间的T分期、M分期、患病年龄、Lauren分型、病理分期和组织学分型等临床特征比较,差异均有统计学意义(P< 0.05)。4个亚型各自有不同的肿瘤标志特征和微环境状态。亚型与特定剪接因子的高表达有关(log2FC > 1且FDR < 0.05),且核心剪接因子的基因集表达与肿瘤的抗原提呈过程有潜在的重要关联(Pearson R= 0.44,P = 0.000)。结论·胃癌可变剪接事件的变化和胃癌的临床表型与肿瘤微环境密切相关。剪接因子的表达水平主导可变剪接水平的变化。剪接因子有望成为胃癌分型的标志物以及改善免疫治疗效果的重要靶点。
中图分类号:
顾琦晟, 张米粒, 曹灿, 李继坤. 基于TCGA数据库分析胃癌可变剪接与肿瘤免疫的关系[J]. 上海交通大学学报(医学版), 2021, 41(4): 448-458.
Qi-sheng GU, Mi-li ZHANG, Can CAO, Ji-kun LI. Association of alternative splicing and tumor immune in gastric cancer based on TCGA data set[J]. JOURNAL OF SHANGHAI JIAOTONG UNIVERSITY (MEDICAL SCIENCE), 2021, 41(4): 448-458.

图1 TCGA中胃癌可变剪切概况Note:A. Histogram of splicing events with values in TCGA-STAD. B.Histogram of standard deviation of PSI value of splicing events in TCGA-STAD. C. Main types of splicing events. D. Association of frequency of splicing events in TCGA-STAD and splicing types.
Fig 1 Overview of alternative splicing of gastric cancer in TCGA
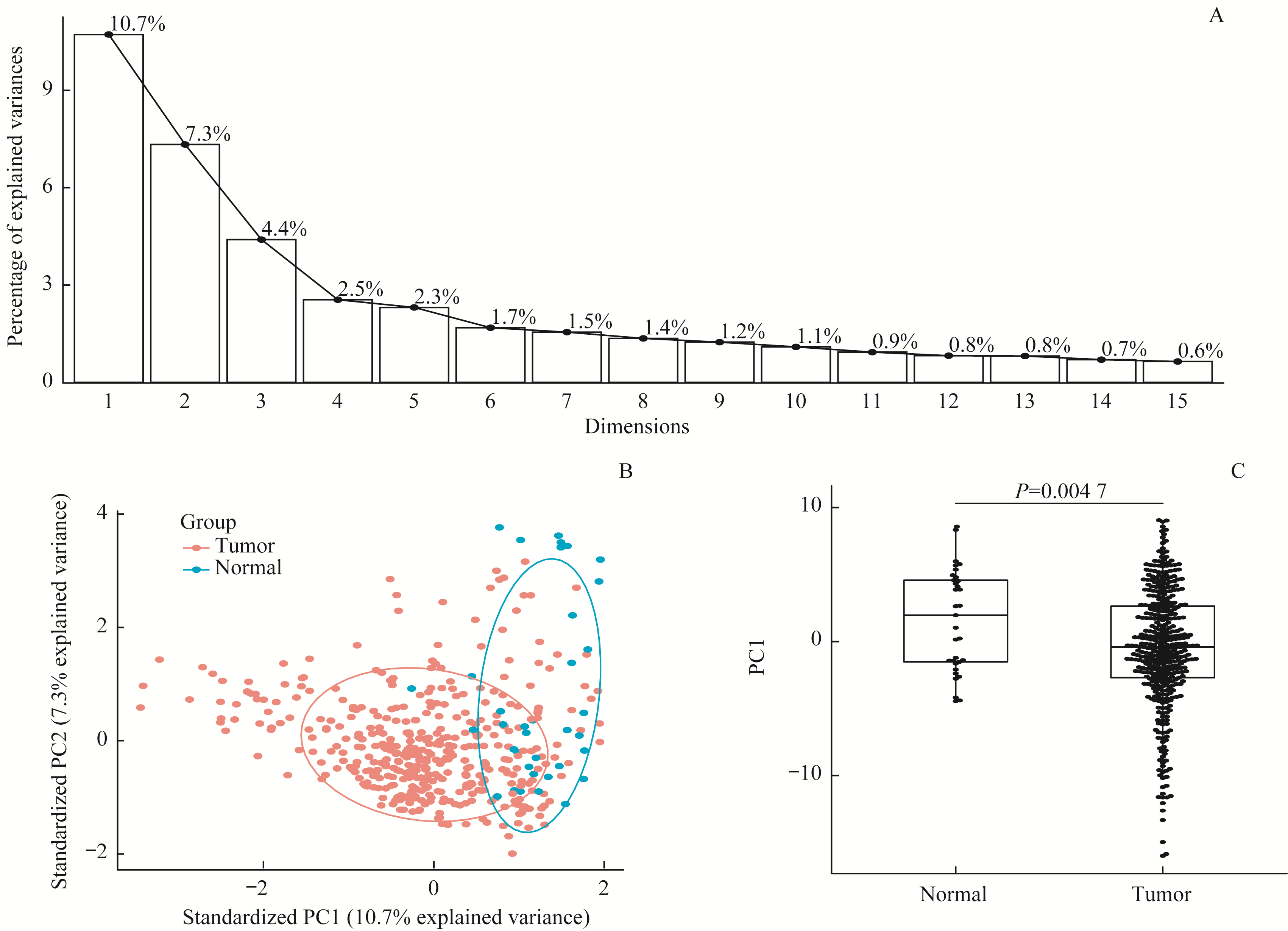
图2 通过PCA对可变剪接事件进行降维分析Note:A.Scree plot shows explained variances of the first ten principle components. B. PCA biplot shows the distribution of splicing events after dimension reduction by PCA. C. Box plot of PC1 value between tumor tissues and adjacent normal tissues.
Fig 2 Dimensional reduction analysis of alternative splicing events by using PCA

图 3 根据可变剪接事件对胃癌样本进行一致性聚类分析Note:A.Correlation matrix heatmaps correspond to cluster number k from 2 to 9. B.Delta area plot indicates the optimal k value. C. PCA biplot of PSI matrix grouped by the 4 subtypes based on consensus clustering.
Fig 3 Consensus clustering of gastric cancer samples based on alternative splicing events
| Item | C1 (n=199) | C2 (n=144) | C3 (n=31) | C4 (n=35) | P value | Missing data/% |
|---|---|---|---|---|---|---|
| Mean age at Initial pathological diagnosis/year | 65.74±10.69 | 66.2±10.20 | 71.13±9.87 | 59.51± 10.47 | 0.000 | 1.2 |
| M stage/n (%) | 0.020 | 5.1 | ||||
| M0 | 181 (96.3) | 125 (91.2) | 24 (82.8) | 33 (97.1) | ||
| M1 | 7 (3.7) | 12 (8.8) | 5 (17.2) | 1 (2.9) | ||
| T stage/n (%) | 0.007 | 2.2 | ||||
| T1 | 18 (9.0) | 3 (2.1) | 1 (3.8) | 0 (0) | ||
| T2 | 30 (15.1) | 41 (29.3) | 9 (34.6) | 8 (22.9) | ||
| T3 | 97 (48.7) | 59 (42.1) | 8 (30.8) | 14 (40.0) | ||
| T4 | 54 (27.1) | 37 (26.4) | 8 (30.8) | 13 (37.1) | ||
| N stage/n (%) | 0.636 | 4.5 | ||||
| N0 | 59 (30.4) | 47 (33.8) | 6 (25.0) | 10 (30.3) | ||
| N1 | 50 (25.8) | 44 (31.7) | 10 (41.7) | 7 (21.2) | ||
| N2 | 43 (22.2) | 22 (15.8) | 5 (20.8) | 8 (24.2) | ||
| N3 | 42 (21.6) | 26 (18.7) | 3 (12.5) | 8 (24.2) | ||
| AJCC pathological stage /n (%) | 0.000 | 5.9 | ||||
| Ⅰ | 30 (15.5) | 19 (14.0) | 5 (22.7) | 3 (8.8) | ||
| Ⅱ | 55 (28.5) | 51 (37.5) | 4 (18.2) | 12 (35.3) | ||
| Ⅲ | 95 (49.2) | 49 (36.0) | 4 (18.2) | 18 (52.9) | ||
| Ⅳ | 13 (6.7) | 17 (12.5) | 9 (40.9) | 1 (2.9) | ||
| Lauren classification /n (%) | 0.000 | 38.6 | ||||
| Diffuse | 36 (25.0) | 16 (23.9) | 3 (23.1) | 21 (77.8) | ||
| Intestinal | 108 (75.0) | 51 (76.1) | 10 (76.9) | 6 (22.2) | ||
| Histological grade/n (%) | 0.000 | 2.2 | ||||
| G1 | 6 (3.1) | 2 (1.4) | 2 (6.5) | 1 (3.1) | ||
| G2 | 87 (44.8) | 51 (35.7) | 9 (29.0) | 1 (3.1) | ||
| G3 | 101 (52.1) | 90 (62.9) | 20 (64.5) | 30 (93.8) | ||
| Gender /n (%) | 0.186 | 0 | ||||
| Female | 60 (30.2) | 54 (37.5) | 14 (45.2) | 15 (42.9) | ||
| Male | 139 (69.8) | 90 (62.5) | 17 (54.8) | 20 (57.1) | ||
| Race /n (%) | 0.045 | 13.7 | ||||
| Asian | 55 (30.1) | 21 (17.2) | 1 (7.7) | 9 (25.7) | ||
| Black | 9 (4.9) | 3 (2.5) | 0 (0) | 0 (0) | ||
| White | 119 (65.0) | 98 (80.3) | 12 (92.3) | 26 (74.3) |
表1 基于可变剪接分类的4种亚型胃癌患者的临床资料比较
Tab1 Comparison of clinical information of patients among the 4 subtypes of gastric cancer based on alternative splicing classification
| Item | C1 (n=199) | C2 (n=144) | C3 (n=31) | C4 (n=35) | P value | Missing data/% |
|---|---|---|---|---|---|---|
| Mean age at Initial pathological diagnosis/year | 65.74±10.69 | 66.2±10.20 | 71.13±9.87 | 59.51± 10.47 | 0.000 | 1.2 |
| M stage/n (%) | 0.020 | 5.1 | ||||
| M0 | 181 (96.3) | 125 (91.2) | 24 (82.8) | 33 (97.1) | ||
| M1 | 7 (3.7) | 12 (8.8) | 5 (17.2) | 1 (2.9) | ||
| T stage/n (%) | 0.007 | 2.2 | ||||
| T1 | 18 (9.0) | 3 (2.1) | 1 (3.8) | 0 (0) | ||
| T2 | 30 (15.1) | 41 (29.3) | 9 (34.6) | 8 (22.9) | ||
| T3 | 97 (48.7) | 59 (42.1) | 8 (30.8) | 14 (40.0) | ||
| T4 | 54 (27.1) | 37 (26.4) | 8 (30.8) | 13 (37.1) | ||
| N stage/n (%) | 0.636 | 4.5 | ||||
| N0 | 59 (30.4) | 47 (33.8) | 6 (25.0) | 10 (30.3) | ||
| N1 | 50 (25.8) | 44 (31.7) | 10 (41.7) | 7 (21.2) | ||
| N2 | 43 (22.2) | 22 (15.8) | 5 (20.8) | 8 (24.2) | ||
| N3 | 42 (21.6) | 26 (18.7) | 3 (12.5) | 8 (24.2) | ||
| AJCC pathological stage /n (%) | 0.000 | 5.9 | ||||
| Ⅰ | 30 (15.5) | 19 (14.0) | 5 (22.7) | 3 (8.8) | ||
| Ⅱ | 55 (28.5) | 51 (37.5) | 4 (18.2) | 12 (35.3) | ||
| Ⅲ | 95 (49.2) | 49 (36.0) | 4 (18.2) | 18 (52.9) | ||
| Ⅳ | 13 (6.7) | 17 (12.5) | 9 (40.9) | 1 (2.9) | ||
| Lauren classification /n (%) | 0.000 | 38.6 | ||||
| Diffuse | 36 (25.0) | 16 (23.9) | 3 (23.1) | 21 (77.8) | ||
| Intestinal | 108 (75.0) | 51 (76.1) | 10 (76.9) | 6 (22.2) | ||
| Histological grade/n (%) | 0.000 | 2.2 | ||||
| G1 | 6 (3.1) | 2 (1.4) | 2 (6.5) | 1 (3.1) | ||
| G2 | 87 (44.8) | 51 (35.7) | 9 (29.0) | 1 (3.1) | ||
| G3 | 101 (52.1) | 90 (62.9) | 20 (64.5) | 30 (93.8) | ||
| Gender /n (%) | 0.186 | 0 | ||||
| Female | 60 (30.2) | 54 (37.5) | 14 (45.2) | 15 (42.9) | ||
| Male | 139 (69.8) | 90 (62.5) | 17 (54.8) | 20 (57.1) | ||
| Race /n (%) | 0.045 | 13.7 | ||||
| Asian | 55 (30.1) | 21 (17.2) | 1 (7.7) | 9 (25.7) | ||
| Black | 9 (4.9) | 3 (2.5) | 0 (0) | 0 (0) | ||
| White | 119 (65.0) | 98 (80.3) | 12 (92.3) | 26 (74.3) |
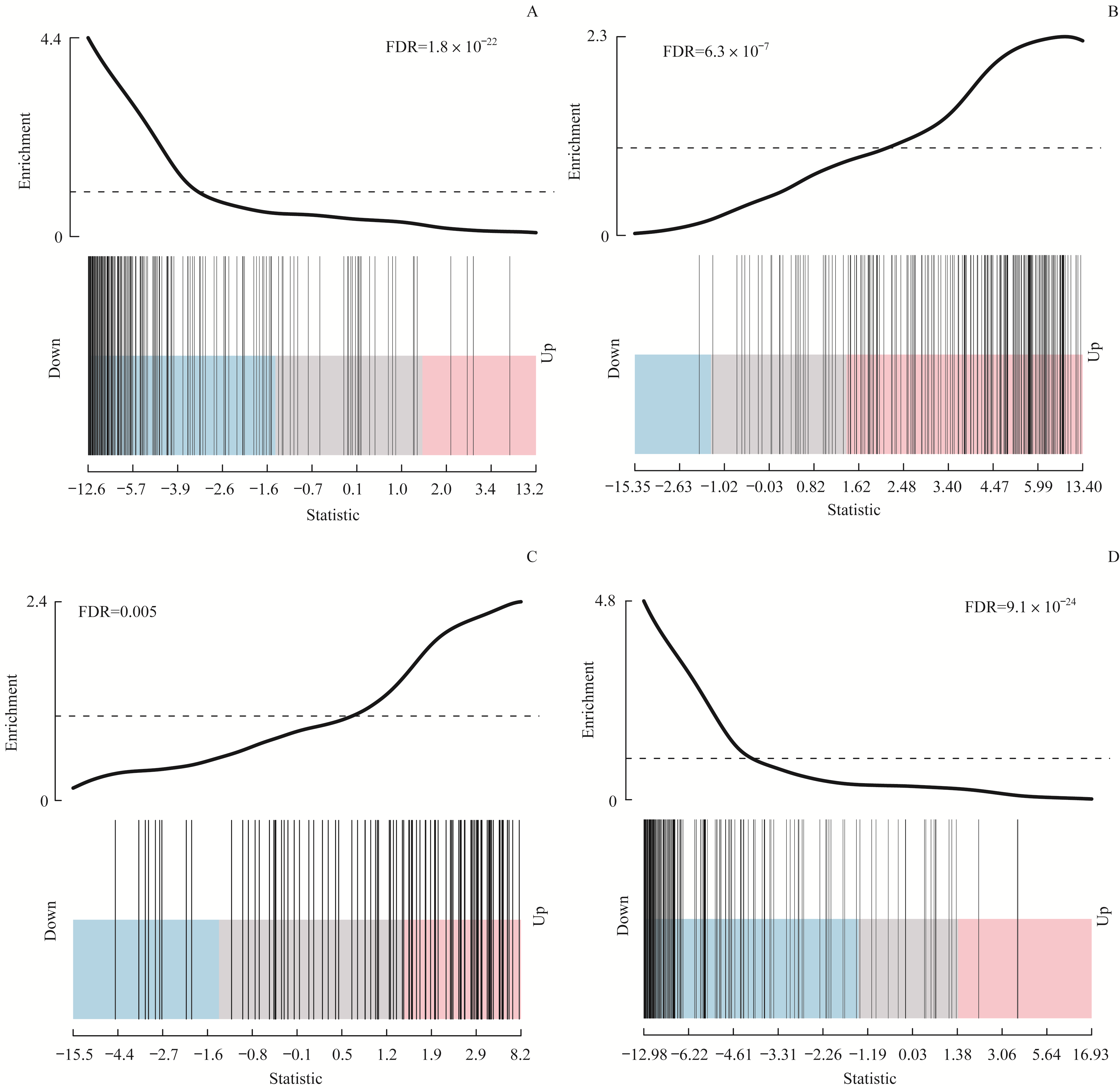
图4 标志基因集在4个可变剪接亚型中的最显著相关通路Note:A. EMT gene set downregulated in C1 type samples. B. EMT gene set upregulated in C2 type samples. C. Protein secretion gene set upregulated in C3 type samples. D. Cell cycle-related targets of E2F downregulated in C4 type samples.
Fig4 The most significantly related pathways of hallmark gene set in the 4 subtypes based on splicing events
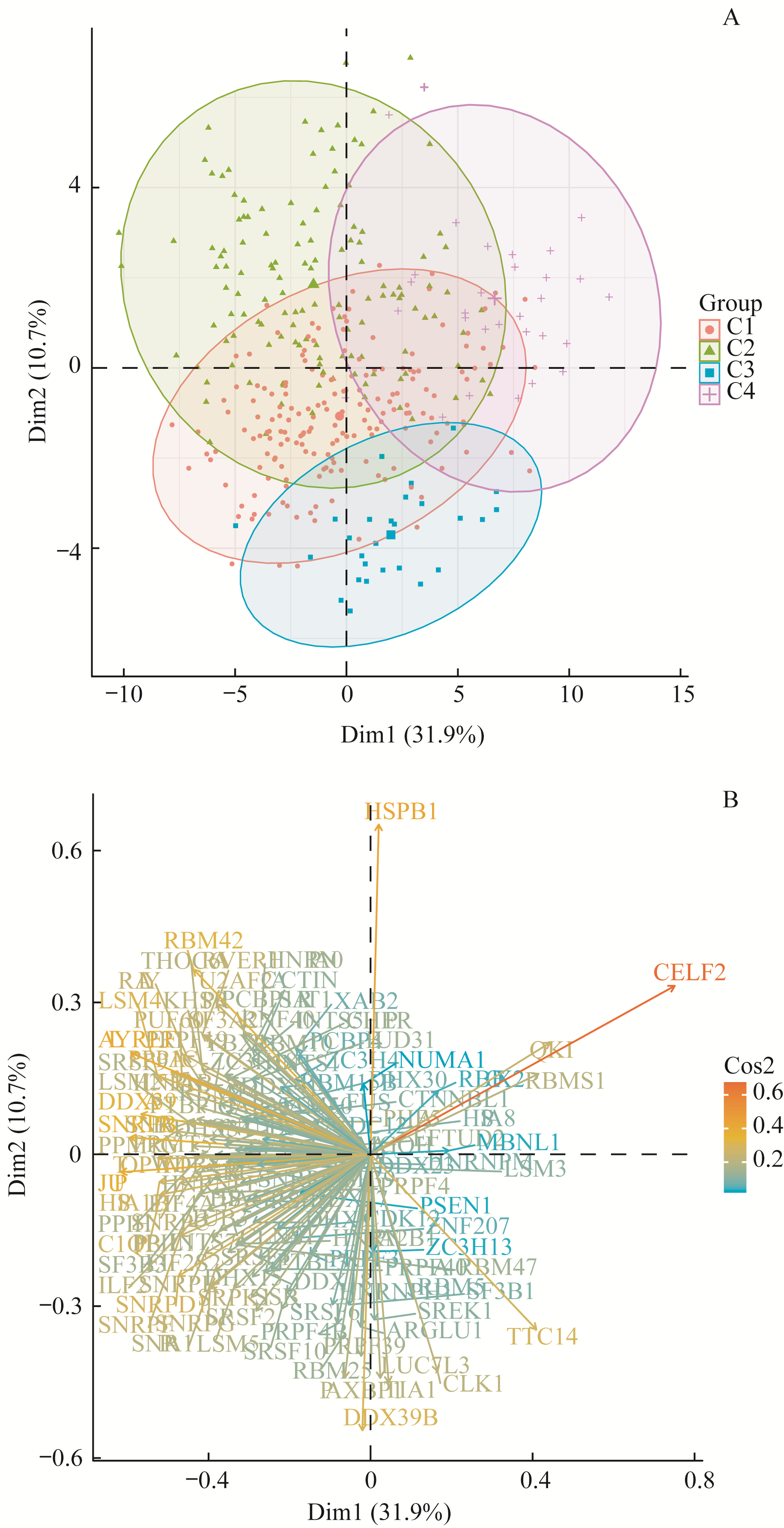
图 7 通过PCA分析可变剪接亚型与剪接因子的关系Note:A. PCA plot shows the clustering of samples grouped by subtypes based on splicing events by using mRNA expression of splicing factors. B. PCA variable plot shows the contribution of main splicing factors in PC1 and PC2.
Fig 7 Association of subtypes based on splicing events and splicing factors using PCA
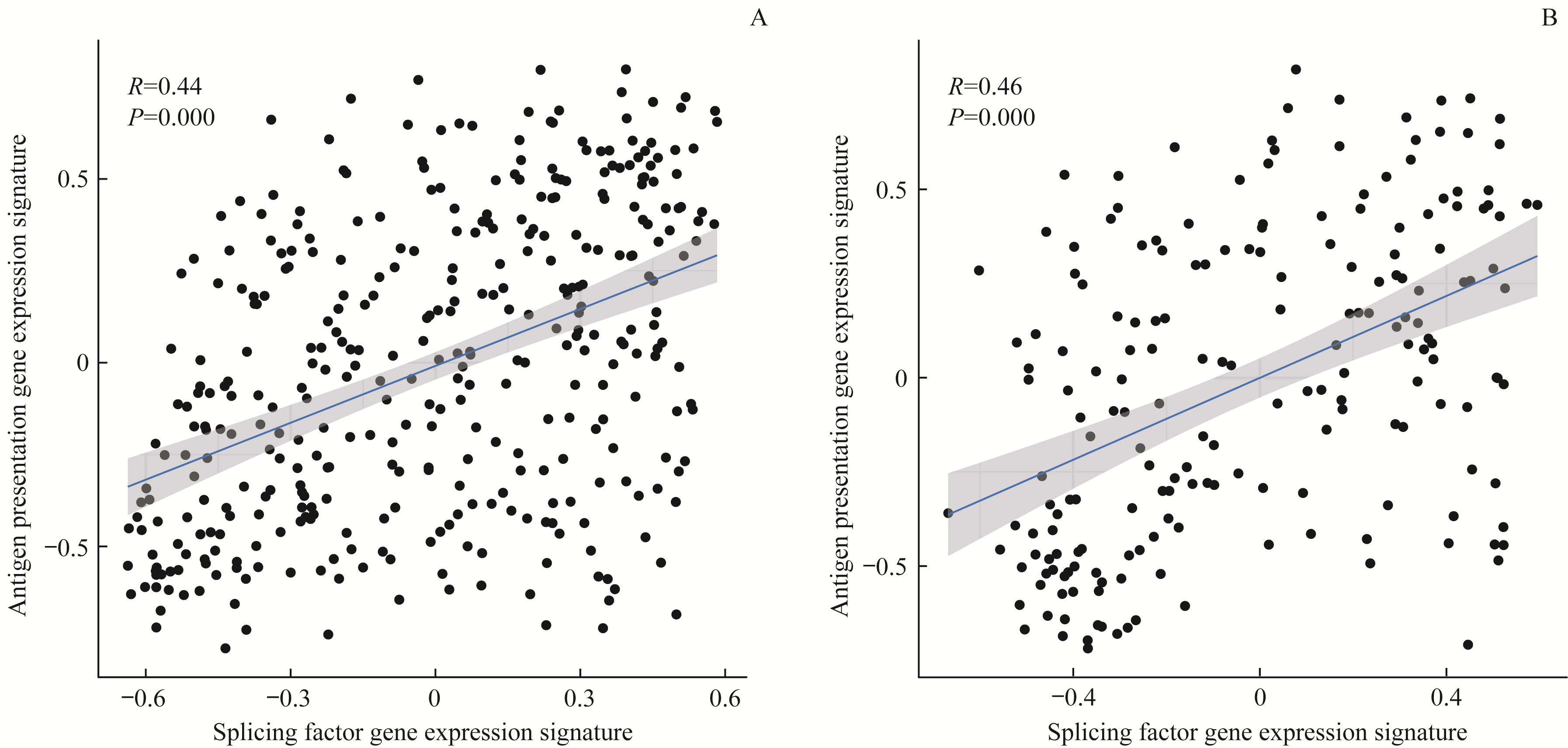
图9 胃癌剪接因子基因表达特征与抗原提呈基因表达特征的关系Note:A. TCGA-STAD. B.GSE15459 gastric cancer samples.
Fig 9 Scatter plots of gene expression signatures between antigen presentation and splicing factors
| 1 | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019[J]. CA Cancer J Clin, 2019, 69(1): 7-34. |
| 2 | Mahoney KM, Freeman GJ, McDermott DF. The next immune-checkpoint inhibitors: pd-1/PD-L1 blockade in melanoma[J]. Clin Ther, 2015, 37(4): 764-782. |
| 3 | Bryan LJ, Gordon LI. Blocking tumor escape in hematologic malignancies: the anti-PD-1 strategy[J]. Blood Rev, 2015, 29(1): 25-32. |
| 4 | Kang YK, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial[J]. Lancet, 2017, 390(10111): 2461-2471. |
| 5 | Stamm S, Ben-Ari S, Rafalska I, et al. Function of alternative splicing [J]. Gene, 2005, 344: 1-20. |
| 6 | Biamonti G, Catillo M, Pignataro D, et al. The alternative splicing side of cancer [J]. Semin Cell Dev Biol, 2014, 32: 30-36. |
| 7 | Kozlovski I, Siegfried Z, Amar-Schwartz A, et al. The role of RNA alternative splicing in regulating cancer metabolism[J]. Hum Genet, 2017, 136(9): 1113-1127. |
| 8 | Ryan M, Wong WC, Brown R, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer[J]. Nucleic Acids Res, 2016, 44(d1): D1018-D1022. |
| 9 | Ooi CH, Ivanova T, Wu J, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer[J]. PLoS Genet, 2009, 5(10): e1000676. |
| 10 | Josse J, Husson F. missMDA: a package for handling missing values in multivariate data analysis [J]. J Stat Softw, 2016, 70(1): 1-31. |
| 11 | Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis [J]. J Stat Softw, 2008, 25(1): 1-18. |
| 12 | Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking[J]. Bioinformatics, 2010, 26(12): 1572-1573. |
| 13 | Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation[J]. Nucleic Acids Res, 2012, 40(17): e133. |
| 14 | Liberzon A, Birger C, Thorvaldsdóttir H, et al. The molecular signatures database hallmark gene set collection [J]. Cell systems, 2015, 1(6): 417-425. |
| 15 | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data [J]. BMC Bioinformatics, 2013, 14(1): 7. |
| 16 | Kahles A, Lehmann KV, Toussaint NC, et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients [J]. Cancer Cell, 2018, 34(2): 211-224. e216. |
| 17 | Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles[J]. NatMethods, 2015, 12(5): 453-457. |
| 18 | Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer[J]. Immunity, 2018, 48(4): 812-830.e14. |
| 19 | Leone P, Shin EC, Perosa F, et al. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells[J]. J Natl Cancer Inst, 2013, 105(16): 1172-1187. |
| 20 | Oltean S, Bates DO. Hallmarks of alternative splicing in cancer[J]. Oncogene, 2014, 33(46): 5311-5318. |
| 21 | Dunn GP, Bruce AT, Ikeda H, et al. Cancer immunoediting: from immunosurveillance to tumor escape[J]. Nat Immunol, 2002, 3(11): 991-998. |
| 22 | Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2[J]. Cell, 1992, 68(2): 365-375. |
| 23 | Venables JP, Lapasset L, Gadea G, et al. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation[J]. Nat Commun, 2013, 4: 2480. |
| 24 | Mallory MJ, Allon SJ, Qiu J, et al. Induced transcription and stability of CELF2 mRNA drives widespread alternative splicing during T-cell signaling[J]. PNAS, 2015, 112(17): E2139-E2148. |
| 25 | Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs[J]. Cell, 2015, 160(6): 1125-1134. |
| 26 | Bian Y, Wang L, Lu H, et al. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis [J]. Biochem Biophys Res Commun, 2012, 422(1): 187-193. |
| [1] | 那迪娜·帕尔哈提, 张鹏善, 徐亦天, 陈赟琪, 黄陈. 人去泛素化酶圆柱瘤蛋白截短体质粒的构建及其对胃癌细胞表型的调控研究[J]. 上海交通大学学报(医学版), 2025, 45(9): 1149-1160. |
| [2] | 杨娜, 刘俊丽, 白静, 杨思怡, 韩继明, 张华华. HENMT1通过激活PI3K-AKT-mTOR信号通路促进胃癌的增殖与迁移[J]. 上海交通大学学报(医学版), 2025, 45(6): 717-726. |
| [3] | 汤开然, 冯成领, 韩邦旻. 基于单细胞测序与转录组测序构建M2巨噬细胞基因相关的前列腺癌预后模型[J]. 上海交通大学学报(医学版), 2025, 45(5): 549-561. |
| [4] | 陈勇羽, 黄益仁, 陈哲逸, 周冰倩, 陈诗宇, 郑英霞. 丝氨酸蛋白酶抑制因子1在胃癌中的表达及其促进胃癌发展的作用机制[J]. 上海交通大学学报(医学版), 2025, 45(2): 150-160. |
| [5] | 张烨晟, 杨易静, 黄依雯, 施珑玙, 王曼媛, 陈思思. 肿瘤微环境免疫细胞调节肿瘤细胞耐药性的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(7): 830-838. |
| [6] | 魏云鑫, 蒋绪顺, 蔡梦瑶, 温睿智, 杜晓刚. COMP与糖尿病肾病自噬相关性分析及其功能验证[J]. 上海交通大学学报(医学版), 2024, 44(7): 847-858. |
| [7] | 刘林楠, 冯莉, 王龙, 刘嘉寅, 范志松. 多能蛋白聚糖在恶性肿瘤中的表达及生物学作用的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(4): 525-530. |
| [8] | 王鑫, 王晓霞, 李彦庆, 郑永鑫, 乌杰, 任猛, 贾向东, 许天祥. 香叶基香叶基焦磷酸合成酶在肺鳞状细胞癌中的表达及临床意义[J]. 上海交通大学学报(医学版), 2024, 44(3): 312-324. |
| [9] | 邓青松, 张长青, 陶诗聪. 烟酰胺代谢相关基因与骨关节炎的关系探索[J]. 上海交通大学学报(医学版), 2024, 44(2): 145-160. |
| [10] | 江爽, 俞继卫. m6A去甲基化酶在胃癌发生发展中的作用机制研究进展[J]. 上海交通大学学报(医学版), 2024, 44(2): 271-277. |
| [11] | 王卫, 王红丽, 阿力比亚提·艾尼, 衣力亚尔·肉苏, 阿依努尔, 杨亮. 血管抑制蛋白2在三阴性乳腺癌中的功能及其调控可变剪接机制[J]. 上海交通大学学报(医学版), 2024, 44(12): 1526-1535. |
| [12] | 张宗稳, 冯莉, 范志松. hnRNPH1在肿瘤生长和转移中作用的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(12): 1615-1620. |
| [13] | 俞思薇, 徐梓淇, 陶梦玉, 范广建. 脂肪酸代谢紊乱通过上调ZNF143促进胰腺癌进展的机制研究[J]. 上海交通大学学报(医学版), 2024, 44(10): 1255-1265. |
| [14] | 李钰, 姜艺凡, 童荣亮, 陈迪宇, 吴健. FOXM1与肿瘤代谢关系的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(10): 1323-1329. |
| [15] | 克德尔亚·艾山江, 傅怡, 赖冬林, 邬海龙, 龚伟. 肝细胞癌相关的核编码线粒体基因及临床信息的综合预后模型[J]. 上海交通大学学报(医学版), 2024, 44(1): 1-12. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||