
Journal of Shanghai Jiao Tong University (Medical Science) ›› 2022, Vol. 42 ›› Issue (11): 1589-1597.doi: 10.3969/j.issn.1674-8115.2022.11.011
• Techniques and methods • Previous Articles Next Articles
SUN Yameng1( ), MA Ye2(
), MA Ye2( ), GUO Runsheng2(
), GUO Runsheng2( )
)
Received:2022-05-07
Accepted:2022-10-18
Online:2022-11-28
Published:2023-01-04
Contact:
MA Ye,GUO Runsheng
E-mail:symbuster_1986@163.com;malele2013@sina.com;grs982600@163.com
Supported by:CLC Number:
SUN Yameng, MA Ye, GUO Runsheng. Detection and analysis of copy number of HER2 gene in breast cancer tissue samples[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2022, 42(11): 1589-1597.
Add to citation manager EndNote|Ris|BibTeX
URL: https://xuebao.shsmu.edu.cn/EN/10.3969/j.issn.1674-8115.2022.11.011
| Gene | Forword primer (5′→3′) | Reverse primer (5′→3′) | Probe sequence (5′→3′) |
|---|---|---|---|
| HER2 | TCACTCATATCCTCCTCTTTCTGC | AATTTTCACATTCTCCCCATCAG | FAM-CAGGGCATCTGGATC-MGB |
| EIF2C1 | GCTGCTAGGCTTTCCTGTTC | GCCTATTTTCCTGCATCTTCT | HEX-AGGCCCCAAAACCCTAAAC-MGB |
Tab 1 List of primers and probe sequences
| Gene | Forword primer (5′→3′) | Reverse primer (5′→3′) | Probe sequence (5′→3′) |
|---|---|---|---|
| HER2 | TCACTCATATCCTCCTCTTTCTGC | AATTTTCACATTCTCCCCATCAG | FAM-CAGGGCATCTGGATC-MGB |
| EIF2C1 | GCTGCTAGGCTTTCCTGTTC | GCCTATTTTCCTGCATCTTCT | HEX-AGGCCCCAAAACCCTAAAC-MGB |
| Sample | Detection System 1 | BC-HER2-2019 | ||||
|---|---|---|---|---|---|---|
| HER2 gene copy number (copies) | Reference gene copy number (copies) | Copy amplification [( HER2/Reference)]×2 | HER2 gene copy number (copies) | Reference gene copy number (copies) | Copy amplification [( HER2/Reference)]×2 | |
| HV-1 | 2 400 | 1.33 | 3 180 | 1.99 | ||
| HV-2 | 2 150 | 1.25 | 3 050 | 1.97 | ||
| HV-3 | 2 600 | 1.41 | 3 200 | 2.05 | ||
Tab 2 Copy number and copy number ratio of HER2 and reference gene in healthy human cfDNA
| Sample | Detection System 1 | BC-HER2-2019 | ||||
|---|---|---|---|---|---|---|
| HER2 gene copy number (copies) | Reference gene copy number (copies) | Copy amplification [( HER2/Reference)]×2 | HER2 gene copy number (copies) | Reference gene copy number (copies) | Copy amplification [( HER2/Reference)]×2 | |
| HV-1 | 2 400 | 1.33 | 3 180 | 1.99 | ||
| HV-2 | 2 150 | 1.25 | 3 050 | 1.97 | ||
| HV-3 | 2 600 | 1.41 | 3 200 | 2.05 | ||
| Sample No. | Concentration/(ng·μL -1) | HER2 Gene copy | Sample No. | Concentration/(ng·μL -1) | HER2 Gene copy |
|---|---|---|---|---|---|
| 1 | 0.44 | 1.80 | 13 | 0.21 | 2.24 |
| 2 | 0.19 | 2.20 | 14 | 0.10 | 2.20 |
| 3 | 0.54 | 2.08 | 15 | 0.16 | 2.22 |
| 4 | 0.26 | 2.01 | 16 | 0.10 | 1.20 |
| 5 | 0.95 | 2.58 | 17 | 0.38 | 2.37 |
| 6 | 0.62 | 2.00 | 18 | 0.13 | 1.87 |
| 7 | 0.23 | 2.90 | 19 | 0.49 | 1.99 |
| 8 | 0.29 | 2.27 | 20 | 0.32 | 1.62 |
| 9 | 0.32 | 1.99 | 21 | 0.10 | 2.40 |
| 10 | 0.21 | 2.28 | 22 | 0.24 | 1.75 |
| 11 | 0.23 | 2.08 | 23 | 0.19 | 2.19 |
| 12 | 0.38 | 2.17 | 24 | 0.74 | 1.69 |
Tab 3 HER2 copy number ratio in 24 healthy human cfDNA samples
| Sample No. | Concentration/(ng·μL -1) | HER2 Gene copy | Sample No. | Concentration/(ng·μL -1) | HER2 Gene copy |
|---|---|---|---|---|---|
| 1 | 0.44 | 1.80 | 13 | 0.21 | 2.24 |
| 2 | 0.19 | 2.20 | 14 | 0.10 | 2.20 |
| 3 | 0.54 | 2.08 | 15 | 0.16 | 2.22 |
| 4 | 0.26 | 2.01 | 16 | 0.10 | 1.20 |
| 5 | 0.95 | 2.58 | 17 | 0.38 | 2.37 |
| 6 | 0.62 | 2.00 | 18 | 0.13 | 1.87 |
| 7 | 0.23 | 2.90 | 19 | 0.49 | 1.99 |
| 8 | 0.29 | 2.27 | 20 | 0.32 | 1.62 |
| 9 | 0.32 | 1.99 | 21 | 0.10 | 2.40 |
| 10 | 0.21 | 2.28 | 22 | 0.24 | 1.75 |
| 11 | 0.23 | 2.08 | 23 | 0.19 | 2.19 |
| 12 | 0.38 | 2.17 | 24 | 0.74 | 1.69 |
| Sample No. | Test date | Copy amplification [( HER2/Reference)]×2 | |||||
|---|---|---|---|---|---|---|---|
| Batch 1 | Batch 2 | Batch 3 | |||||
| Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | ||
| 1 | Day 1 | 1.74 | 2.14 | 2.16 | 2.14 | 1.82 | 1.94 |
| 2 | Day 2 | 1.84 | 1.88 | 2.36 | 2.18 | 1.92 | 1.88 |
| 3 | Day 3 | 2.08 | 1.98 | 2.38 | 2.50 | 2.20 | 1.82 |
| 4 | Day 4 | 2.12 | 2.26 | 1.83 | 2.14 | 1.80 | 1.90 |
| 5 | Day 5 | 2.10 | 2.10 | 2.24 | 2.04 | 2.18 | 1.92 |
| Xmean | 2.05 | ||||||
| Intra batch standard deviation | 0.14 | ||||||
| inter batch standard deviation | 0.19 | ||||||
| Intra batch precision/% | 6.80 | ||||||
| inter batch precision/% | 9.40 | ||||||
Tab 4 Precision evaluation of the detection system
| Sample No. | Test date | Copy amplification [( HER2/Reference)]×2 | |||||
|---|---|---|---|---|---|---|---|
| Batch 1 | Batch 2 | Batch 3 | |||||
| Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | ||
| 1 | Day 1 | 1.74 | 2.14 | 2.16 | 2.14 | 1.82 | 1.94 |
| 2 | Day 2 | 1.84 | 1.88 | 2.36 | 2.18 | 1.92 | 1.88 |
| 3 | Day 3 | 2.08 | 1.98 | 2.38 | 2.50 | 2.20 | 1.82 |
| 4 | Day 4 | 2.12 | 2.26 | 1.83 | 2.14 | 1.80 | 1.90 |
| 5 | Day 5 | 2.10 | 2.10 | 2.24 | 2.04 | 2.18 | 1.92 |
| Xmean | 2.05 | ||||||
| Intra batch standard deviation | 0.14 | ||||||
| inter batch standard deviation | 0.19 | ||||||
| Intra batch precision/% | 6.80 | ||||||
| inter batch precision/% | 9.40 | ||||||
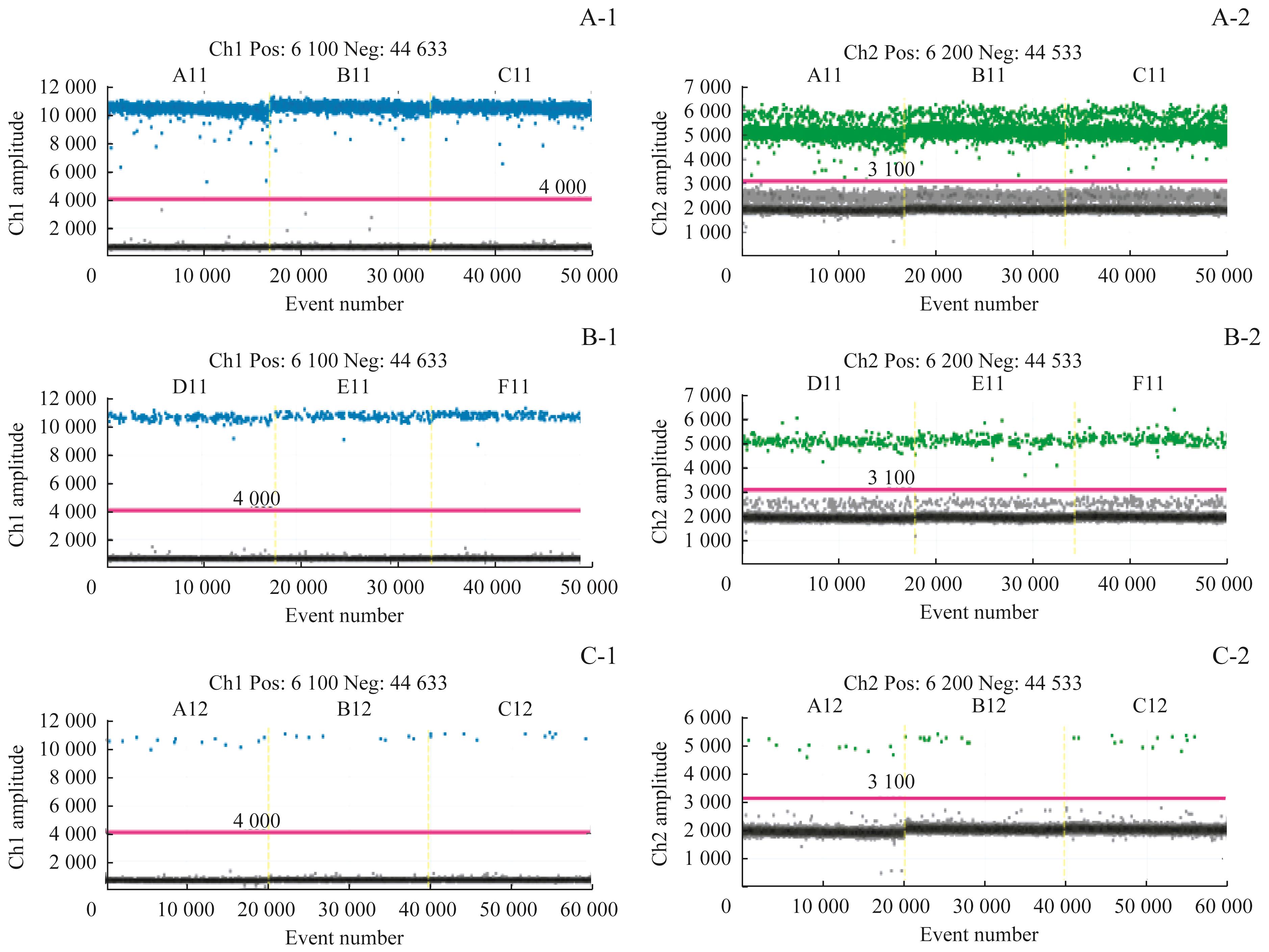
Fig 2 Results of sensitivity detectionNote: A-1/A-2. Results of three duplication holes of HER2 and reference gene by using 10ng. B-1/B-2. Results of three duplication holes of HER2 and reference gene by using 1ng. C-1/C-2. Results of three duplication holes of HER2 and reference gene by using 0.1ng.
| Loading quantity | Repeat | HER2 Gene copy number (copies/20 μL) | Reference Gene copy number (copies/20 μL) | Copy amplification [( HER2/Reference)]×2 | CV/% |
|---|---|---|---|---|---|
| 10 ng | 1 | 2 780 | 1.89 | 3.30 | |
| 2 | 3 100 | 1.99 | |||
| 3 | 3 160 | 2.01 | |||
| 1 ng | 1 | 226 | 274 | 1.64 | 12.50 |
| 2 | 234 | 244 | 1.91 | ||
| 3 | 276 | 260 | 2.11 | ||
| 0.1 ng | 1 | 15.20 | 14.00 | 2.20 | 26.00 |
| 2 | 9.60 | 14.40 | 1.30 | ||
| 3 | 13.00 | 15.40 | 1.70 |
Tab 5 Copy number statistics and CV of sensitivity detection
| Loading quantity | Repeat | HER2 Gene copy number (copies/20 μL) | Reference Gene copy number (copies/20 μL) | Copy amplification [( HER2/Reference)]×2 | CV/% |
|---|---|---|---|---|---|
| 10 ng | 1 | 2 780 | 1.89 | 3.30 | |
| 2 | 3 100 | 1.99 | |||
| 3 | 3 160 | 2.01 | |||
| 1 ng | 1 | 226 | 274 | 1.64 | 12.50 |
| 2 | 234 | 244 | 1.91 | ||
| 3 | 276 | 260 | 2.11 | ||
| 0.1 ng | 1 | 15.20 | 14.00 | 2.20 | 26.00 |
| 2 | 9.60 | 14.40 | 1.30 | ||
| 3 | 13.00 | 15.40 | 1.70 |
HER2∶ reference | Theoretical value | Actual value | |||||
|---|---|---|---|---|---|---|---|
HER2 gene copy number (copies/20 μL) | Reference gene copy number (copies/20 μL) | Copy amplification [( HER2/ reference)]×2 | HER2 gene copy number (copies/20 μL) | Reference gene copy number (copies/20 μL) | Copy amplification [( HER2/ reference)]×2 | ||
| 1∶1 | 3 260 | 2.04 | 3 160 | 1.89 | |||
| 2∶1 | 5 984 | 3.93 | 6 460 | 2.99 | |||
| 3∶1 | 8 708 | 6.01 | 8 900 | 3 000 | 5.94 | ||
| 4∶1 | 13 914 | 7.96 | 16 140 | 8.62 | |||
| 5∶1 | 14 482 | 2927 | 9.90 | 16 760 | 10.00 | ||
Tab 6 Copy number statistics of linear detection
HER2∶ reference | Theoretical value | Actual value | |||||
|---|---|---|---|---|---|---|---|
HER2 gene copy number (copies/20 μL) | Reference gene copy number (copies/20 μL) | Copy amplification [( HER2/ reference)]×2 | HER2 gene copy number (copies/20 μL) | Reference gene copy number (copies/20 μL) | Copy amplification [( HER2/ reference)]×2 | ||
| 1∶1 | 3 260 | 2.04 | 3 160 | 1.89 | |||
| 2∶1 | 5 984 | 3.93 | 6 460 | 2.99 | |||
| 3∶1 | 8 708 | 6.01 | 8 900 | 3 000 | 5.94 | ||
| 4∶1 | 13 914 | 7.96 | 16 140 | 8.62 | |||
| 5∶1 | 14 482 | 2927 | 9.90 | 16 760 | 10.00 | ||
| No. | NGS copy amplification | ddPCR copy amplification |
|---|---|---|
| 1 a | 1.90 | 1.62 |
| 2 a | 1.90 | 1.86 |
| 3 a | 1.90 | 1.62 |
| 4 a | 5.00 | 4.27 |
| 5 a | 6.00 | 4.21 |
| 6 a | 13.10 | 14.10 |
| 7 a | 8.00 | 6.64 |
| 8 a | 18.00 | 16.00 |
| 9 a | 2.40 | 1.73 |
| 9 b | 2.10 | 1.84 |
| 10 a | 2.20 | 1.77 |
| 10 b | 1.80 | 1.41 |
| 11 a | 5.00 | 3.50 |
| 11 b | 1.80 | 1.49 |
| 12 a | 5.00 | 3.85 |
| 12 b | 1.70 | 1.20 |
Tab 7 Copy number results by NGS comparison of ddPCR
| No. | NGS copy amplification | ddPCR copy amplification |
|---|---|---|
| 1 a | 1.90 | 1.62 |
| 2 a | 1.90 | 1.86 |
| 3 a | 1.90 | 1.62 |
| 4 a | 5.00 | 4.27 |
| 5 a | 6.00 | 4.21 |
| 6 a | 13.10 | 14.10 |
| 7 a | 8.00 | 6.64 |
| 8 a | 18.00 | 16.00 |
| 9 a | 2.40 | 1.73 |
| 9 b | 2.10 | 1.84 |
| 10 a | 2.20 | 1.77 |
| 10 b | 1.80 | 1.41 |
| 11 a | 5.00 | 3.50 |
| 11 b | 1.80 | 1.49 |
| 12 a | 5.00 | 3.85 |
| 12 b | 1.70 | 1.20 |
| ddPCR | FISH | Specificity [% (95% CI)] | Sensitivity [% (95% CI)] | Kappa value | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Positive | 22 | 12 | 76.5 (62.2—86.8) | 84.6 (64.3—95.0) | 0.57 |
| Negative | 4 | 39 | |||
Tab 8 Gold standard FISH to evaluate the HER2 copy number detected by ddPCR
| ddPCR | FISH | Specificity [% (95% CI)] | Sensitivity [% (95% CI)] | Kappa value | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Positive | 22 | 12 | 76.5 (62.2—86.8) | 84.6 (64.3—95.0) | 0.57 |
| Negative | 4 | 39 | |||
| 1 | PONDÉ N, AFTIMOS P, PICCART M. Antibody-drug conjugates in breast cancer: a comprehensive review[J]. Curr Treat Options Oncol, 2019, 20(5): 37. |
| 2 | GRIGUOLO G, PASCUAL T, DIECI M V, et al. Interaction of host immunity with HER2-targeted treatment and tumor heterogeneity in HER2-positive breast cancer[J]. J Immunother Cancer, 2019, 7(1): 90. |
| 3 | MAHTANI R, HOLMES F A, BADVE S, et al. Breast Cancer Therapy Expert Group (BCTEG). A roundtable discussion of the Breast Cancer Therapy Expert Group (BCTEG): clinical developments and practice guidance on human epidermal growth factor receptor 2 ( HER2)-positive breast Cancer[J]. Clin Breast Cancer, 2020, 20(3): e251-e260. |
| 4 | WOLFF A C, HAMMOND M E H, ALLISON K H, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update[J]. J Clin Oncol, 2018, 36(20): 2105- 2122. |
| 5 | NIU D F, LI L, YU Y, et al. Evaluation of next generation sequencing for detecting HER2 copy number in breast and gastric cancers[J]. Pathol Oncol Res, 2020, 6(4): 2577-2585. |
| 6 | AHN S, WOO J W, LEE K, et al. HER2 status in breast cancer: changes in guidelines and complicating factors for interpretation[J]. J Pathol Transl Med, 2020, 54(1): 34-44. |
| 7 | 相学平. CEP17异常对评判乳腺癌 HER2基因扩增状态的影响[J]. 临床与实验病理学杂志, 2014, 30(7): 728-731. |
| XIANG X P. Effects of CEP17 abnormal on HER2 gene status evaluation in breast carcinoma[J]. Chin J Clin Exp, 2014, 30(7): 728-731. | |
| 8 | 卢仁泉, 柳光宇, 杨文涛, 等. 外周血 HER2基因扩增检测(数字PCR法)在抗 HER2治疗中的应用共识[J]. 中国癌症杂志, 2022, 32(1): 90-96. |
| LU R Q, LIU G Y, YANG W T, et al. Application consensus of peripheral blood HER2 gene amplification detection (digital PCR) in anti- HER2 therapy[J]. China Oncol, 2022, 32(1): 90-96. | |
| 9 | SHODA K, ICHIKAWA D, FUJITA Y, et al. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer[J]. Gastric Cancer, 2017, 20(1): 126-135. |
| 10 | SHODA K, MASUDA K, ICHIKAWA D, et al. HER2 amplification detected in the circulating DNA of patients with gastric cancer: a retrospective pilot study[J]. Gastric Cancer, 2015, 18(4):698-710. |
| 11 | ZHU Y Z, LU D, LIRA M E, et al. Droplet digital polymerase chain reaction detection of HER2 amplification in formalin fixed paraffin embedded breast and gastric carcinoma samples[J]. Exp Mol Pathol, 2016, 100(2): 287-293. |
| 12 | 杨文涛,步宏.乳腺癌 HER2检测指南(2019版)[J].中华病理学杂志, 2019, 48(3): 169-175. |
| YANG W T, BU H. Guidelines for detection of HER2 in breast cancer (2019 edition)[J]. Chin J Pathol, 2019, 48(3): 169-175. | |
| 13 | 陈静瑶, 周杰, 李飞, 等. 飞燕草素通过AKT/mTOR通路诱导HER-2+乳腺癌细胞自噬[J]. 中南大学学报(医学版), 2017, 42(3): 264-270. |
| CHEN S Y, ZHOU J, LI F, et al. Delphinidin induces autophagy in HER-2+ breast cancer cells via inhibition of AKT/mTOR pathway[J]. J Cent South Univ (Med Sci), 2017, 42(3): 264-270. | |
| 14 | 董周寰, 张晶, 王哲,等. 运用数字PCR检测乳腺癌组织FFPE样品中人表皮生长因子受体2拷贝数的变化[J]. 生物技术通讯, 2018, 29(3): 6. |
| DONG Z H, ZHANG J, WANG Z, et al. HER2 Copy Number Alteration in FFPE Samples Using Droplet Digital PCR[J]. Lett Biotechnol, 2018, 29(3): 6. | |
| 15 | BARTLEY A N, WASHINGTON M K, COLASACCO C, et al. HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: guideline from the College of American Pathologists, American Society for Clinical Pathology, and the American Society of Clinical Oncology [J]. J Clin Oncol, 2017, 35(4): 446-464. |
| 16 | JANJIGIAN Y Y, KAWAZOE A, YAÑEZ P, et al. The KEYNOTE-811 trial of dual PD-1 and HER2 blockade in HER2-positive gastric cancer[J]. Nature,2021, 600(7890): 727-730. |
| 17 | 曹明丽. 基于分子分型的乳腺癌流行病学新认识[J]. 中国肿瘤临床, 2017, 44(9): 449-451. |
| CAO M L. Advances in breast cancer epidemiology based on molecular subtyping[J]. Chin J Clin Oncol, 2017, 44(9): 449-451. | |
| 18 | 张艳秋, 王昳凡, 王简. HER-2阳性乳腺癌的新辅助治疗现状及展望[J]. 临床肿瘤学杂志, 2017, 22(3): 264-271. |
| ZHANG Y Q, WANG Y F, WANG J. The current and progress of neoadjuvant treatment of HER-2 positive breast cancer[J]. Chin Clin Oncol, 2017, 22(3): 264-271. | |
| 19 | VESCI L, CAROLLO V, ROSCILLI G, et al. Trastuzumab and docetaxel in a preclinical organotypic breast cancer model using tissue slices from mammary fat pad: translational relevance[J]. Oncol Rep. 2015, 34(3): 1146-1152. |
| 20 | 李明, 王涛, 杨周. 靶向药物在HER2阳性乳腺癌患者化疗中的临床应用价值研究[J]. 中国医学创新, 2022, 19(8): 51-54. |
| LI M, WANG T, YANG Z. The clinical application research of targeted drugs in chemotherapy of HER2 positive breast cancer patients[J]. Chinese Med Innovations, 2022, 19(8): 51-54. | |
| 21 | 王利锋, 袁芳, 陈锐, 等. 微滴数字PCR在检测乳腺癌 HER2基因扩增中的应用[J]. 中华病理学杂志, 2018, 47(10): 790-792. |
| WANG L F, YUAN F, CHEN R, et al. Application of droplet digital PCR technology for HER2 gene amplification of breast cancer[J]. Chin J Pathol, 2018, 47(10): 790-792. | |
| 22 | KIM B, NAM S K, SEO S H, et al. Comparative analysis of HER2 copy number between plasma and tissue samples in gastric cancer using droplet digital PCR[J]. Sci Rep, 2020, 10(1): 4177. |
| 23 | WANG Y, TSANG J Y S, CUI Y, et al. Robust and accurate digital measurement for HER2 amplification in HER2 equivocal breast cancer diagnosis[J]. Sci Rep, 2017, 7(1): 6752. |
| 24 | WANG H, LI B, LIU Z, et al. HER2 copy number of circulating tumor DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer[J]. Eur J Cancer, 2018, 88:92-100. |
| 25 | LEE K S, NAM S K, SEO S H, et al. Digital polymerase chain reaction for detecting c-MYC copy number gain in tissue and cell-free plasma samples of colorectal cancer patients[J]. Sci Rep, 2019, 9(1): 1611. |
| [1] | WANG Jingyi, DENG Jiali, ZHU Yi, DING Xinyi, GUO Jiajing, WANG Zhongling. Experimental study on novel pH-responsive manganese-based nanoprobes for ferroptosis and magnetic resonance imaging in breast cancer [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(9): 1183-1193. |
| [2] | LI Long, ZHAO Xia, JIN Shan, LI Zeying, LÜ Fuqiang, PANG Lijuan, LIU Kejian. Deciphering the protective role of AZGP1 in heart failure through Mendelian randomization [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(8): 1035-1045. |
| [3] | LIN Tong, TAO Yi, JIN Shiwei, SUN Miao, MI Jianqing. Effect of plasma exchange combined with classical chemotherapy on renal function in patients with multiple myeloma [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(7): 823-828. |
| [4] | SU Xinglei, LU Ping, PENG Junjie, WANG Zimin, SONG Ping, HAN Da. Comparison of DNA and RNA extraction efficiency from blood [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(4): 476-486. |
| [5] | DENG Jiali, GUO Jiajing, WANG Jingyi, DING Xinyi, ZHU Yi, WANG Zhongling. Self -assembled drug -loaded nanoprobes for pyroptosis sensitization and chemical exchange saturation transfer imaging in breast cancer [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(3): 271-281. |
| [6] | WU Shiyi, CHEN Si, LIU Bohan, LIU Yuting, LIU Yiwen, HE Yiqing, DU Yan, ZHANG Guoliang, GUO Qian, GAO Feng, YANG Cuixia. Role of "HA coat" in modulating stemness and endocrine resistance in ER+ breast cancer [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(10): 1298-1307. |
| [7] | WU Qizhen, LIU Qiming, CHAI Yezi, TAO Zhengyu, WANG Yinan, GUO Xinning, JIANG Meng, PU Jun. Evaluation of machine learning prediction of altered inflammatory metabolic state after neoadjuvant therapy for breast cancer [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(9): 1169-1181. |
| [8] | HAN Yishan, XU Ziqi, TAO Mengyu, FAN Guangjian, YU Bo. PRMT6 promotes the proliferation and migration of breast cancer cells [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(8): 999-1010. |
| [9] | LIU Yiqing, JIN Chenxi, FENG Baoyi, CHENG Zhenzhe, SUN Yilin, ZHENG Xiaofei, DONG Tingting, WU Hao, TAO Yong. Atp2b2 Oblivion heterozygous mutation causes progressive vestibular dysfunction in mice [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(6): 723-732. |
| [10] | WANG Wei, WANG Hongli, ALIBIYATI·i Ain, YILIYAER· Rousu, AYI NUER, YANG Liang. Function of vasohibin-2 and the mechanism of alternative splicing in triple-negative breast cancer [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(12): 1526-1535. |
| [11] | TAN Chen, XU Zhangrun, XUE Yang, CHEN Jiayu, YAO Lijun. Research progress in drug repurposing in the treatment of breast cancer [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(11): 1454-1459. |
| [12] | DU Shaoqian, TAO Mengyu, CAO Yuan, WANG Hongxia, HU Xiaoqu, FAN Guangjian, ZANG Lijuan. CXCL9 expression in breast cancer and its correlation with the characteristics of tumor immunoinfiltration [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(7): 860-872. |
| [13] | CAO Yuan, WANG Hongxia, ZHU Ying, LI Junjian. Expression of tetraspanin 1 in breast cancer and its mechanism in promoting the progression of breast cancer [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(3): 293-300. |
| [14] | YANG Xiaoxuan, ZHU Shan, QIAN Cheng, CHU Xiaoying. Effect of intraoperative use of low-dose dexmedetomidine on the prognosis of patients undergoing breast cancer surgery [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(2): 194-200. |
| [15] | WANG Lanxi, MA Guanrong, JIANG Yongzhu, CHANG Xiulin, FANG Liaoqiong, BAI Jin. Effects of Escherichia coli outer membrane vesicles on proliferation of breast cancer cells and tumor growth of tumor-bearing mice [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(10): 1245-1254. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||