干扰素调节因子3(interferon regulatory factor 3,IRF3)是与干扰素基因密切相关的细胞因子,以单体形式表达于多种细胞中,功能与病毒感染等有关[4]。IRF3需要通过病毒感染等诱导其C端结构的磷酸化进而转移进入细胞核内,最后与相关功能转录因子结合从而激活干扰素的分泌途径,这一过程在先天性机体免疫抗感染中占据重要地位[5]。但近期研究[6-10]发现,IRF3在某些恶性肿瘤组织中高表达且与患者的预后相关,这说明IRF3可能在恶性肿瘤发生发展的过程发挥作用。已有报道提示,病毒感染或双链RNA(double-stranded RNA,dsRNA)激活IRF3,是通过作用于IRF3蛋白C端末端(第385~405个氨基酸)实现的;其中7个关键位点(第385/386/396/398/402/404/405个氨基酸)的苏氨酸或丝氨酸(threonine/serine)残基簇的磷酸化被证明是IRF3发挥功能活性所必需的[11-12]。将IRF3-396/398/402/404/405(D)(即IRF3-5D)中的苏氨酸或丝氨酸残基突变为丙氨酸(alanine)或天冬氨酸(asparticacid)时,发现其可被进一步结合或激活;然而IRF3-385/386(D)未被检测到明显的功能活性,说明这2个位点突变为天冬氨酸的拟磷酸化形式不足以发挥IRF3磷酸化激活后的功能,并且IRF3-385/386位点残基被丙氨酸取代则会阻止信号的激活,因此385/386位点被认为是IRF3功能激活的核心区域[10-13]。本研究通过探索IRF3的核心激活区域的相关功能,探讨IRF3在结直肠癌恶性进程中可能发挥的作用。通过体外实验,本研究分析高表达IRF3-5D蛋白的肠癌细胞在肿瘤恶性增殖和侵袭行为上的变化,以及IRF3激活后是否调控一些肿瘤相关蛋白在转录水平上的表达,希冀研究结果为结直肠癌等恶性肿瘤的靶向免疫治疗提供潜在治疗靶点,并为结直肠恶性肿瘤的防治提供更多研究方向和理论基础。
1 材料与方法
1.1 数据下载与临床标本收集
从癌症基因组图谱(The Cancer Genome Atlas,TCGA)数据库获取原发性肾癌、结直肠癌、肝癌和前列腺癌患者生存随访数据与相应癌组织、癌旁组织IRF3蛋白表达水平检测结果。数据来源于877例原发性肾癌、597例原发性结直肠癌、365例原发性肝癌和494例原发性前列腺癌患者。选取2022年在上海交通大学医学院附属新华医院行恶性肿瘤切除术的肾癌或者结直肠癌患者共10例,相关组织切片由新华医院病理科提供。
1.2 细胞、质粒、主要试剂与仪器
人源肾细胞系HEK-293T,结直肠癌细胞系CT26、COLON26购自中国科学院上海生命科学院典型培养物保藏委员会细胞库。细胞系通过实验室培养验证确定。所有细胞系均无异常污染。质粒pLX302、pcDNA3购自美国Addgene公司。IRF3-WT质粒序列由中国上海生命科学院郎靖瑜课题组提供。
抗程序性死亡配体1(programmed death-ligand 1,PD-L1)抗体、抗IRF3抗体、抗磷酸化IRF3抗体(phosphorylated IRF3,p-IRF3)抗体、抗IRF9抗体、抗信号转导和转录激活子2(signal transducer and activator of transcription 2,STAT2)抗体、抗甘油醛-3-磷酸脱氢酶(glyceraldehyde-3-phosphate dehydrogenase,GAPDH)抗体购自美国CST公司。DMEM/F-12培养基、胰酶、青-链霉素双抗购自美国Gibco公司。4%多聚甲醛、0.05%结晶紫染液购自江苏碧云天公司。脱脂奶粉、5%牛血白蛋白、ECL发光液购自上海生工生物工程股份有限公司。嘌呤霉素购自上海雅酶生物科技有限公司。TRAF家族相关的NF-κB激活因子(TRAF family member-associated NF-κB activator,TANK)结合激酶1(TANK-binding kinase 1,TBK1)抑制剂BX795(T1830)购自美国TargetMol公司。
倒置荧光电子显微镜购自上海尼康仪器有限公司,低温水平离心机购自德国Eppendorf公司,荧光定量PCR仪购自美国ABI公司,细胞培养箱购自美国Themo公司。
1.3 免疫组织化学法
将患者肾癌或者结直肠癌病理组织置入去蜡液中静置15 min,加入无水乙醇洗涤,而后使用1×磷酸盐缓冲液(phosphate buffered saline,PBS;pH 7.5)洗涤,切片。5%H2O2溶液孵育30 min后用1×PBS(pH 7.5)洗涤。3%牛血白蛋白覆盖组织30 min。IRF3一抗(工作浓度1∶1 000)4 ℃孵育过夜后,二抗(工作浓度1∶1 000)室温孵育1 h;1×PBS洗涤3次后,使用3-氨基-9-乙基咔唑(3-amino-9-ethylcarbazole,AEC)显色液孵育30 min,加水终止染色。苏木精染核3 min,加水终止,加入苏木素分化液10 s后流水冲洗。置于光学显微镜下进行结果判读。
1.4 质粒的构建与细胞转染
设计并获取突变型质粒IRF3-5D(396/398/402/404/405-D)、IRF3-2A5D(385/386-A;396/398/402/404/405-D)对应引物序列。配制聚合酶链式反应(polymerase chain reaction,PCR)反应体系,包括引物(10 µmoL)1 µL、高保真PCR Mix预混液(2×)5 µL、TDNA gene-DNA 200 ng,加双蒸馏水至10 µL。PCR仪设置反应程序:95 ℃变性5 min;95 ℃变性30 s,50 ℃~70 ℃设置5个温度梯度,各退火30 s,72 ℃条件延伸30 s,30个循环;72 ℃条件延伸10 min。将目的基因片段与载体质粒双侧酶切形成黏性末端连接后,取2 µL连接产物采用感受肽转化于大肠埃希菌,使用氨苄抗性培养基筛选并获取目标质粒。将质粒(IRF3-WT、IRF3-5D、IRF3-2A5D)通过磷酸钙转染法表达于HEK-293T细胞。取部分转染后12、24 h的HEK-293T细胞培养基内加入TBK1抑制剂BX795(工作浓度为100 µmol/L)。取部分转染后的HEK-293T细胞继续分别培养48和72 h。
1.5 蛋白质印迹法
取“1.4”经TBK1抑制剂处理的HEK-293T细胞,采用细胞裂解法提取细胞蛋白,二喹啉甲酸(bicinchoninic acid,BSA)法统一SDS聚丙烯酰胺凝胶电泳(SDS polyacrylamide gel electrophoresis,SDS-PAGE)上样蛋白浓度。湿转法转膜,按照标准蛋白标记物大小指示裁剪、截取硝酸纤维素膜。将洗净后的蛋白条带置于5%脱脂奶粉封闭液或者5%牛血白蛋白封闭液内,室温下孵育1 h,而后条带上滴加待检测蛋白的特异性一抗(工作浓度1∶1 000),于4 ℃孵育过夜;二抗(工作浓度1∶1 000)室温条件下孵育目标条带2 h;洗膜后完全浸入发光液,设置程序固定曝光时间点与次数,保存显影图像;实验重复3次。
1.6 RNA测序
取“1.4”转染后继续分别培养48、72 h的HEK-293T细胞的60%样本进行转录本测序检测,获得3组(6个亚组:IRF3-WT48 h、IRF3-WT72 h、IRF3-5D48 h、IRF3-5D72 h、IRF3-2A5D48 h、IRF3-2A5D72 h)RNA测序(RNA sequencing,RNA-seq)数据,并进行基因本体(Gene Ontology,GO)功能富集分析与京都基因和基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)通路分析。分别使用GO条目与KEGG路径前20个P值最低者进行映射作图。
1.7 慢病毒包装、细胞感染及蛋白表达检测
将“1.4”中获得载体模板为pLX-302的目标质粒(IRF3-5D、IRF3-WT)通过慢病毒转染法分别感染并表达于结肠癌细胞CT26、COLON26。慢病毒转染前将细胞CT26、COLON26(1×106个)分别铺至培养皿中。培养24 h后,用含有6 μg/mL嘌呤毒素的8 mL新鲜DMEM/F-12培养基替换原培养基,分别加入1 mL(分别含IRF3-5D、IRF3-WT或者空白对照Vector)的病毒悬液。继续培养24 h后,用新鲜培养基替换含有病毒的培养基,继续培养48 h。得到过表达不同蛋白IRF3的3种CT26细胞(CT26IRF3-5D、CT26IRF3-WT、CT26Vector)和3种COLON26细胞(COLON26Vector、COLON26IRF3-WT和COLON26IRF3-5D)。采用蛋白质印迹法验证相关蛋白表达水平。
1.8 细胞计数法
收获生长状态良好的3种CT26细胞(CT26IRF3-5D、CT26IRF3-WT、CT26Vector)和3种COLON26细胞(COLON26Vector、COLON26IRF3-WT和COLON26IRF3-5D)分别制成细胞悬液并计数。将每个培养皿分别铺下5×104个/mL的细胞进行培养。24 h后,记录细胞计数值,而后每24 h计数1次。根据细胞计数结果,绘制细胞生长曲线。
1.9 细胞划痕实验
取3种CT26细胞(CT26IRF3-5D、CT26IRF3-WT、CT26Vector),分别在干净的6孔板中按每2 mL DMEM/F12培养基加入大约5×105个细胞,轻轻摇动混匀后放置于培养箱内培养24 h,至细胞密度约为90%。第2日用直尺辅助,保持枪头与6孔板板底垂直做划痕。用1 mL PBS溶液清洗细胞3次,去除所有漂浮的细胞,然后加入2 mL不含胎牛血清的DMEM/F12培养基继续培养。分别于划痕后的24和48 h观察并拍照。通过测量不同时间点的划痕间距并计算差值,对细胞的迁移能力作出判断。
1.10 克隆形成实验
取3种COLON26细胞(COLON26Vector、COLON26IRF3-WT和COLON26IRF3-5D),对细胞进行分选培养。以每孔500或者2 000个细胞的梯度密度,分别将细胞铺在含有37 ℃预热培养液的6孔板中。恒温37 ℃培养箱孵育,每周更换1次培养基,14 d后取出,弃去上清液,用适量PBS小心清洗2次。加1 mL多聚甲醛室温固定细胞20 min。吸出固定液,加适量0.05%结晶紫室温染色30 min,然后用流水缓慢洗去染液,倒扣晾干。在显微镜下计算有10个以上细胞的克隆数。计算克隆形成率=(克隆数/接种细胞数)×100%。
1.11 统计学分析
数据收集后使用GraphPad Prim 8软件进行统计分析。患者蛋白表达水平与生存分析数据采用单因素方差法分析并作图,体外细胞分子数据通过单因素方差分析或非参数Kruskal-Wallis检验计算组间差异。P<0.05表示差异具有统计学意义。
2 结果
2.1 IRF3的表达水平与患者预后的关系
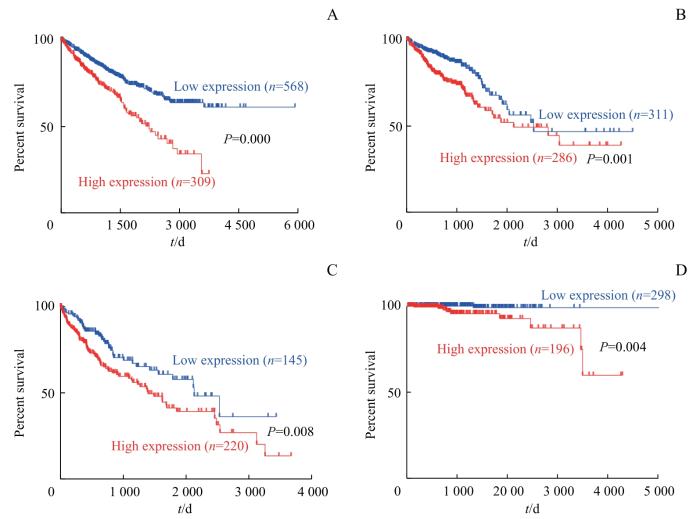
采用TCGA数据库中的不同癌症患者及相关癌组织IRF3蛋白表达数据进行生存分析,结果如图1所示,IRF3表达水平与癌症患者(肾癌、结直肠癌、肝癌和前列腺癌)预后不良密切相关,IRF3表达水平高的患者20年生存率明显下降。
图1
图1
基于不同IRF3表达水平的Kaplan-Meier生存分析
Fig 1
Kaplan-Meier survival analysis based on different IRF3 expression levels
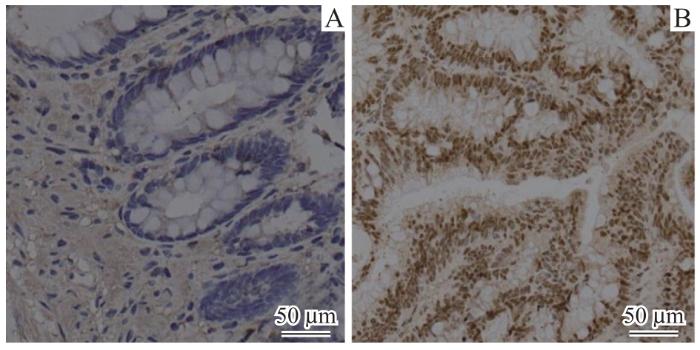
2.2 IRF3蛋白在肾癌和结直肠癌组织中核内高表达
图2
图2
肠癌患者癌组织与癌旁正常组织IRF3蛋白免疫组织化学对比(×20)
Fig 2
Comparison of immunohistochemical staining for IRF3 protein between cancerous and normal tissues adjacent to cancer in patients with colorectal cancer (×20)
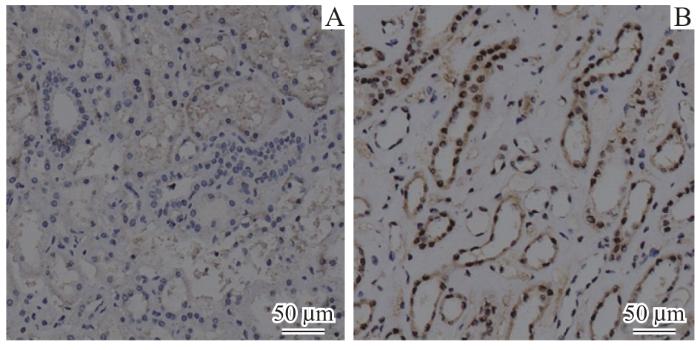
图3
图3
肾癌患者癌组织与癌旁正常组织的IRF3免疫组织化学对比(×20)
Fig 3
Comparison of immunohistochemical staining for IRF3 protein between cancerous and normal tissues adjacent to cancer from patients with renal cancer (×20)
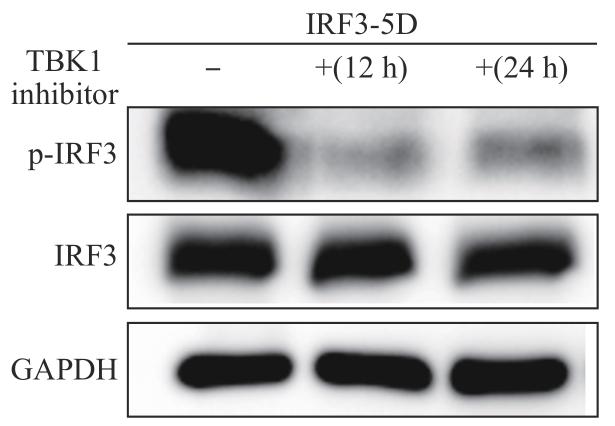
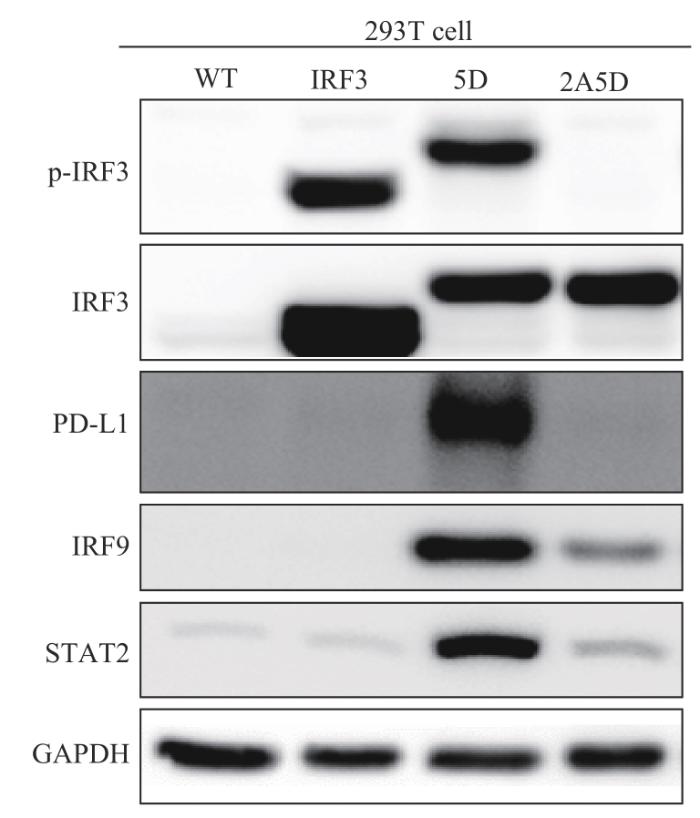
2.3 拟磷酸化IRF3-5D激活依赖TBK1介导的通路
在HEK-293T细胞中过表达拟磷酸化IRF3-5D,分别在12、24 h培养时间点使用TBK1抑制剂后,检测IRF3/p-IRF3(Ser386)蛋白表达水平的变化,结果(图4)发现,p-IRF3(Ser386)表达水平明显下降。
图4
图4
不同时间点下使用TBK1抑制剂后IRF3表达变化
Fig 4
Changes in IRF3 expression with TBK1 inhibitor at different time points
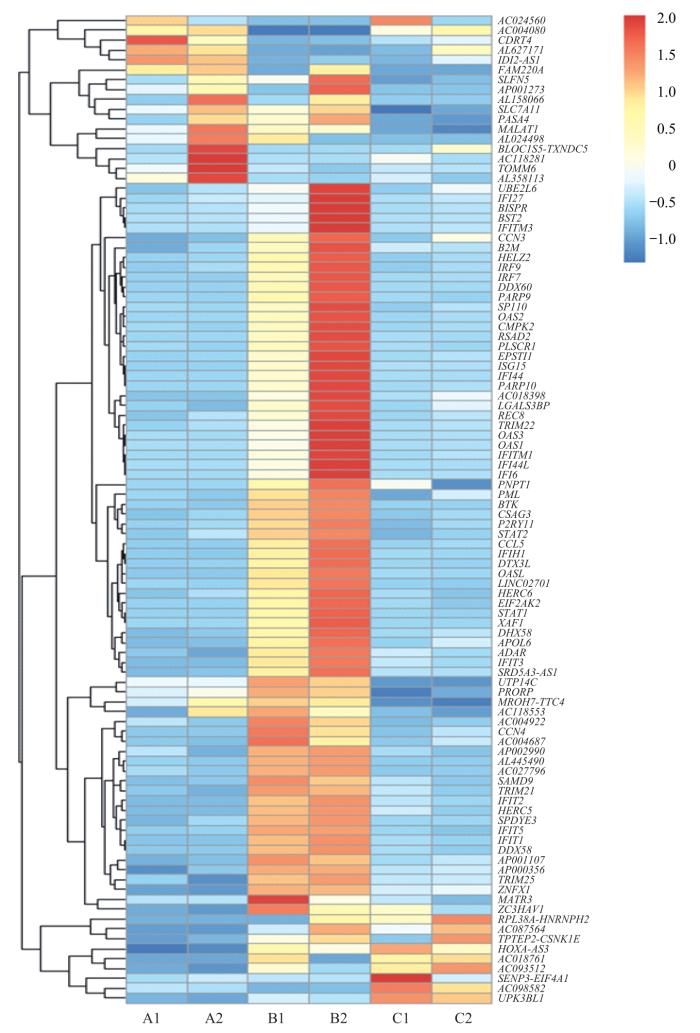
2.4 IRF3-WT/5D/2A5D转染细胞后于转录水平上探索差异表达基因
IRF3-WT/5D/2A5D高表达于HEK-293T细胞系,培养至48 h或者72 h。采用RNA-seq获得基因相对表达数据,探索3组间基因差异化表达情况(图5)。
图5
图5
3组间差异基因聚类热图
Note: Horizontal coordinates are experimental groups. A1: IRF3-WT48 h, A2: IRF3-WT72 h; B1: IRF3-5D48 h, B2: IRF3-5D72 h; C1: IRF3-2A5D48 h, C2: IRF3-2A5D72 h. Vertical coordinates are the names of the major differential genes detected at the transcriptional level. Blue via white to red indicates low to high expression; red indicates high expression genes, and blue indicates low expression genes. CDRT4—CMT1A duplicated region transcript 4; SLFN5—recombinant Schlafen family member 5; RASA4—RAS p21 protein activator 4; MALAT-1—metastasis associated in lung denocarcinoma transcript 1; TOMM6—translocase of outer mitochondrial membrane 6; BISPR—BST2 interferon-stimulated positive regulator; IFITM3—interferon-induced transmembrane protein 3; CCN3—cellular communication network factor 3; B2M—β-2 microglobulin; HELZ2—helicase with zinc finger 2; IRF9—interferon regulatory factor 9; IRF7—interferon regulatory factor 7; DDX60—DExD/H-box helicase 60; PARP9—poly (ADP-ribose) polymerase family member 9; SP110—SP110 nuclear body protein; OAS2—2′-5′-oligoadenylate synthetase 2; CMPK2—cytidine monophosphate kinase 2; RSAD2—radical S-adenosyl methionine domain containing 2; IFI27—interferon alpha-inducible protein 27; BST-2—bone marrow stromal antigen 2; PLSCR1—phospholipid scramblase 1; EPSTI1—epithelial stromal interaction 1; PARP10—poly (ADP-ribose) polymerase family member 10; REC8—REC8 meiotic recombination protein; TRIM22—tripartite motif containing 22; PNPT1—polyribonucleotide nucleotidyltransferase 1; BTK—Bruton tyrosine kinase; STAT2—signal transducer and activator of transcription 2; DTX3L—deltex E3 ubiquitin ligase 3L; HERC6—HECT and RLD domain containing E3 ubiquitin protein ligase family member 6; XAF1—XIAP-associated factor 1; DHX58—DExH-box helicase 58; APOL6—apolipoprotein L6; PRORP—protein-only RNase P catalytic subunit; CCN4—cellular communication network factor 4; SAMD9L—sterile α motif domain containing 9 like; TRIM21—tripartite-motif protein 21; DHX58—DExH-box helicase 58; ZNFX1—zinc finger NFX1-type containing 1; MATR3—matrin 3.
Fig 5
Heat map of differential gene clusterings between the three groups
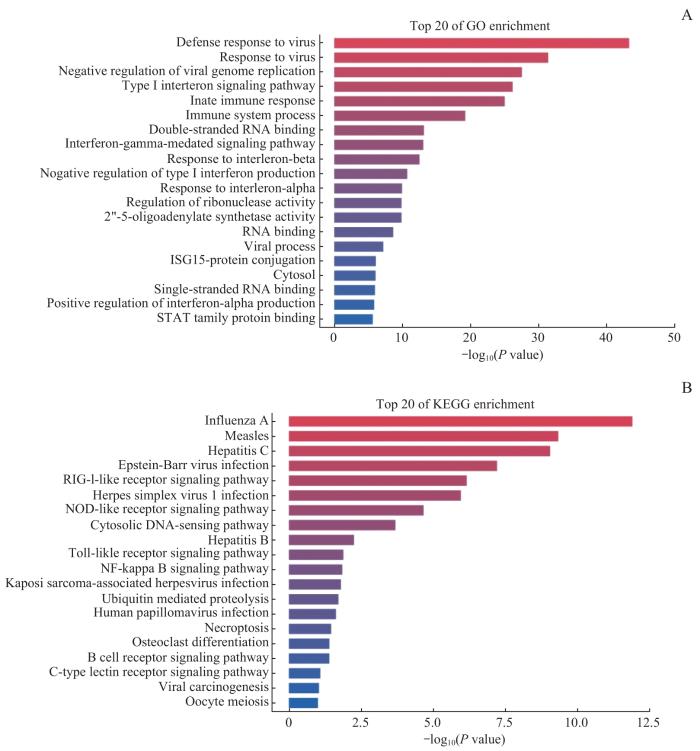
对上述3组间差异表达基因进行GO与KEGG富集分析,展示相关差异表达基因的主要生物功能以及所处的通路信息。结果(图6)显示,IRF3-5D高表达组中,传统干扰素调节与转录翻译等功能通路涉及基因表达水平上调。因此,在IRF3-5D高表达条件下,排除传统研究中干扰素相关基因条目,一些肿瘤相关基因,如BST2、CMPK2、DTX3L、IRF9、SP110、REC8等表达水平明显上调。
图6
图6
GO(A)、KEGG(B)基因富集分析图
Fig 6
Gene enrichment analysis graphs of GO(A) and KEGG(B)
2.5 IRF3-5D可以上调细胞肿瘤相关蛋白的表达水平
在HEK-293T细胞中分别过表达3种IRF3蛋白(IRF3-WT、IRF3-5D、IRF3-2A5D),并检测下游调控蛋白IRF9的表达情况。结果(图7)显示,IRF3-5D高表达组内,IRF9蛋白的表达水平显著上调,与转录水平IRF9的基因表达变化趋势一致;与肿瘤密切相关的PD-L1和STAT2的蛋白表达变化也与IRF9的表达变化趋势具有相似性。
图7
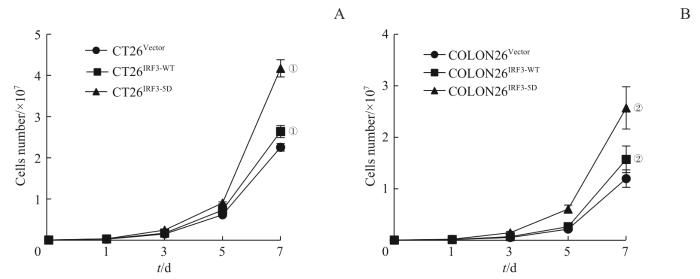
2.6 CT26、COLON26细胞系高表达拟磷酸化蛋白IRF3-5D后对结直肠恶性肿瘤细胞恶性增殖水平的促进作用
将IRF3-5D、IRF3-WT过表达于肠癌细胞后,细胞计数结果(图8)显示:高表达IRF3-5D蛋白的CT26和COLON26细胞表现出更高水平的增殖速度与增殖活力。
图8
图8
3组CT26、COLON26细胞增殖能力对比
Note: ①P=0.000, compared with CT26Vector; ②P=0.000, compared with COLON26Vector.
Fig 8
Comparison of growing capacity among three groups of CT26 and COLON26 cell
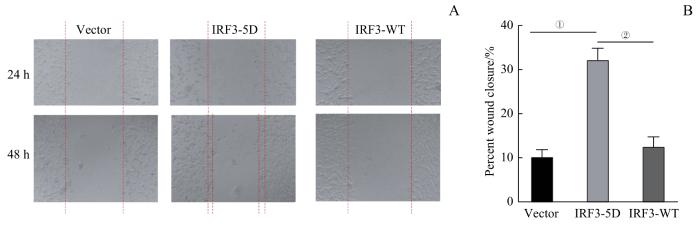
2.7 CT26、COLON26高表达IRF3-5D后提高结直肠恶性肿瘤细胞转移侵袭及生长活力
采用不同蛋白过表达的3种CT26细胞(CT26IRF3-5D、CT26IRF3-WT、CT26Vector)进行细胞划痕实验,结果(图9)显示:CT26IRF3-5D细胞相较于CT26Vector和CT26IRF3-WT细胞,侵袭迁移范围更为明显(均P=0.000)。
图9
图9
3组CT26细胞转移侵袭能力对比
Note: A. Detection of migration capacity of CT26 cells. B. Statistical analysis of CT26 cell wound closure percentage. ①P=0.000, compared between IRF3-5D and
Fig 9
Comparison of invasive ability among three groups of CT26 cells
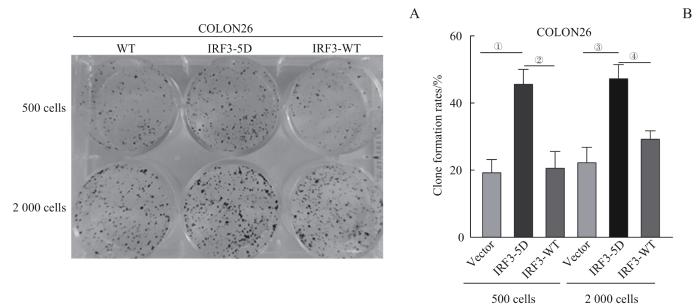
取COLON26Vector、COLON26IRF3-WT和COLON26IRF3-5D细胞进行克隆增殖实验,结果(图10)显示:COLON26IRF3-5D相较于COLON26Vector和COLON26IRF3-WT,克隆增殖更为密集(均P=0.001)。
图10
图10
3组COLON26细胞生长活力对比
Note: Initial cell number laid down was 500 and 2 000. A. Colony formation of COLON26 cells. B. Statistical analysis of clone formation rate of COLON26 cells. ①P=0.000, compared between IRF3-5D and Vector; ②P=0.000, compared between IRF3-5D and IRF3-WT; ③P=0.001, compared between IRF3-5D and Vector; ④P=0.001, compared between IRF3-5D and IRF3-WT.
Fig 10
Comparison of cell viability among three groups of COLON26 cells
3 讨论
结直肠癌是世界上最常见的恶性肿瘤之一。目前,对于结直肠癌的最佳防治策略仍然是提高筛查率,努力实现肠恶性肿瘤的早发现、早诊断与早治疗,从而提高患者的生存率。然而约60%的患者仍以晚期癌症确诊,其中25%~40%的患者癌症持续进展,最终发展为转移性难治性恶性肿瘤[14]。针对晚期结直肠恶性肿瘤的治疗,过去数年里经历了从单纯应用化学药物,到与分子靶向药物和免疫药物结合的转变。探索和发现新的治疗靶点是生物医学研究人员最为紧迫的任务之一。
目前在IRF3激活与其生物学功能相关研究中,IRF3蛋白的激活功能结构域成为研究的重点。IRF3点突变功能实验验证了其激活的关键区域位于C端的氨基酸残基(第385~405位点);同时,通过观察体内激活实验现象,提出了TBK1可能通过作用于IRF3上的2个特殊启动子区域直接或间接激活IRF3的假设,但是目前未于体外实验中完成分子水平的验证[13]。基于当前的研究[21-24]成果,本研究构建了拟磷酸化IRF3模型:将IRF3蛋白分子中第396/398/402/404/405位点的苏氨酸/丝氨酸残基替换成天冬氨酸,随后采用TBK1激酶介导第385/386位点的丝氨酸残基的特异性磷酸化来实现IRF3的招募与活化。
本研究通过体外实验观察得出,在蛋白水平上,拟磷酸化IRF3-5D高表达的细胞中,p-IRF3(Ser386)表达水平的上调同样受到TBK1介导调节。这一现象与既往报道的IRF3依赖TBK1非典型激活通路理论具一致性[25]。因此,体内与体外实验均提示,在IRF3功能通路中,TBK1可能直接或者间接参与IRF3磷酸化过程。
为了进一步探索IRF3-5D拟磷酸化蛋白可能发挥的生物学调控功能,我们将IRF3-WT/5D/2A5D分别高表达于细胞后行RNA-seq,以探索3组细胞的基因差异化表达情况。基因富集分析结果提示细胞内BST2、CMPK2、DTX3L等基因表达水平与IRF3-5D表达水平呈正相关。大量研究[26-34]已报道这些蛋白的高表达与多种癌症的临床预后不良相关。同时,我们发现在IRF3-5D蛋白高表达细胞内,PD-L1、STAT2、IRF9蛋白的表达水平也显著上调,很多研究[35-39]也证明了这一点。因此,细胞中IRF3-5D高表达可能显著增加这些促癌蛋白的表达水平,从而导致IRF3过表达在结直肠癌细胞中的促恶性进展的生物学行为。
相较于CT26Vector、COLON26Vector、COLON26IRF3-WT、CT26IRF3-WT细胞,高表达IRF3-5D蛋白的结直肠癌细胞COLON26IRF3-5D、CT26IRF3-5D均呈现较高的细胞增殖活力,肿瘤细胞增殖速度显著上升、侵袭迁移能力更为明显。因此,IRF3-5D蛋白可能与原发性结直肠癌中表达水平提高的IRF3蛋白具有相似的生物学功能与作用机制。
综上,本研究对于IRF3-5D蛋白相关功能的探索为结直肠癌的潜在治疗靶点研究提供了基础。然而,具体下游作用分子及调控机制尚未完全明确,仍有待进一步研究。
作者贡献声明
徐文晖参与实验设计、实验操作、数据分析、论文写作及修改;杨畅、李瑞卿参与实验操作;卞京、李夏伊、宋梦晴参与资料获取与分析;郑磊贞负责实验指导与课题资助。所有作者均阅读并同意了最终稿件的提交。
AUTHOR's CONTRIBUTIONS
XU Wenhui participated in the experimental design, experimental manipulation, data analysis, paper writing and revision. YANG Chang and LI Ruiqing participated in the experimental manipulation. BIAN Jing, LI Xiayi, and SONG Mengqing participated in the data acquisition and analysis. ZHENG Leizhen was responsible for the experimental guidance and project funding. All authors have read the final manuscript and agreed to the submission.
利益冲突声明
所有作者声明不存在利益冲突。
COMPETING INTERESTS
All authors disclose no relevant conflict of interests.
参考文献