
上海交通大学学报(医学版) ›› 2023, Vol. 43 ›› Issue (5): 545-559.doi: 10.3969/j.issn.1674-8115.2023.05.005
梅艳青1( ), 韩雨洁1, 翁文筠1, 张蕾1,2, 唐玉杰1,2(
), 韩雨洁1, 翁文筠1, 张蕾1,2, 唐玉杰1,2( )
)
收稿日期:2023-03-08
接受日期:2023-04-10
出版日期:2023-05-28
发布日期:2023-07-11
通讯作者:
唐玉杰,电子信箱:yujietang@shsmu.edu.cn。作者简介:梅艳青(1998—),女,硕士生;电子信箱:yanqingmei@sjtu.edu.cn。
基金资助:
MEI Yanqing1( ), HAN Yujie1, WENG Wenyun1, ZHANG Lei1,2, TANG Yujie1,2(
), HAN Yujie1, WENG Wenyun1, ZHANG Lei1,2, TANG Yujie1,2( )
)
Received:2023-03-08
Accepted:2023-04-10
Online:2023-05-28
Published:2023-07-11
Contact:
TANG Yujie, E-mail: yujietang@shsmu.edu.cn.Supported by:摘要:
目的·筛选高级别胶质瘤(high-grade gliomas,HGGs)共有的表观转录靶向治疗新策略,并进行体外治疗效果的测试与相关分子机制的探究。方法·对HGGs中恶性程度和致死率均较高的多个胶质母细胞瘤(glioblastoma,GBM)和弥漫性内生性脑桥胶质瘤(diffuse intrinsic pontine glioma,DIPG)细胞系进行表观转录相关的靶向小分子药物库筛选和基于CRISPR-Cas9系统的功能基因组筛选,以寻找在GBM和DIPG中共同的表观转录靶向治疗新策略。然后针对筛选得到的目标表观转录调控因子,分别测试经CRISPR-Cas9方法敲除该基因以及对应的靶向小分子处理对GBM和DIPG细胞系的体外生长、细胞增殖与凋亡的影响。继而对靶向小分子处理的GBM和DIPG细胞系进行RNA-seq转录组分析,解析其抗肿瘤分子机制。基于此分析结果,通过实时荧光定量PCR(quantitative real-time PCR,RT-qPCR)、蛋白质印迹法以及流式细胞术进一步验证目标表观转录调控因子的抗肿瘤分子机制。结果·针对表观转录调控因子的靶向小分子药物库筛选和功能基因组筛选鉴定出CDK12和CDK13(CDK12/13)是GBM和DIPG共同的潜在治疗新靶点。在多个GBM和DIPG细胞系中,通过CRISPR-Cas9方法敲除CDK12能够显著降低其体外细胞活性。CDK12/13抑制剂SR-4835或THZ531也均能够通过拮抗细胞增殖和促进细胞凋亡造成这2类HGGs细胞系的体外生长受到显著抑制。SR-4835处理GBM和DIPG细胞之后的RNA-seq转录组分析结果表明,HGGs细胞中受CDK12/13抑制剂作用而表达显著下调的基因主要富集在转录调控、DNA损伤反应(DNA damage response,DDR)通路、泛素-蛋白酶体通路以及细胞周期等生物学过程。进一步通过一系列实验方法验证了在DIPG和GBM中,靶向抑制CDK12/13显著下调DDR相关基因的转录进而引起DNA损伤的累积,并诱导细胞发生G2-M细胞周期阻滞。结论·CDK12/13是GBM和DIPG这2类HGGs共同的潜在表观转录靶向治疗靶标;该发现为后续体内模型验证、组合治疗测试提供了理论支持,也为未来进一步的临床转化应用奠定了基础。
中图分类号:
梅艳青, 韩雨洁, 翁文筠, 张蕾, 唐玉杰. 靶向抑制CDK12/13在高级别胶质瘤中的体外治疗效果和作用分子机制探究[J]. 上海交通大学学报(医学版), 2023, 43(5): 545-559.
MEI Yanqing, HAN Yujie, WENG Wenyun, ZHANG Lei, TANG Yujie. In vitro therapeutic effects and molecular mechanisms of targeted inhibition of CDK12/13 in high-grade gliomas[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(5): 545-559.
| Primer | Sequence (5'→3') |
|---|---|
| sgGFP forward | CACCGGGGCGAGGAGCTGTTCACCG |
| sgGFP reverse | AAACCGGTGAACAGCTCCTCGCCCC |
| sgCDK12-2 forward | CACCGACTGACCGACTGCCTTCT |
| sgCDK12-2 reverse | AAACCGAGAAGGCAGTCGGTCAGTC |
| sgCDK12-3 forward | CACCGATTCACCAGTTCAGTATCTG |
| sgCDK12-3 reverse | AAACCAGATACTGAACTGGTGAATC |
表1 sgRNA oligo序列
Tab 1 Oligo sequences of sgRNA
| Primer | Sequence (5'→3') |
|---|---|
| sgGFP forward | CACCGGGGCGAGGAGCTGTTCACCG |
| sgGFP reverse | AAACCGGTGAACAGCTCCTCGCCCC |
| sgCDK12-2 forward | CACCGACTGACCGACTGCCTTCT |
| sgCDK12-2 reverse | AAACCGAGAAGGCAGTCGGTCAGTC |
| sgCDK12-3 forward | CACCGATTCACCAGTTCAGTATCTG |
| sgCDK12-3 reverse | AAACCAGATACTGAACTGGTGAATC |
| Primer | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| ATM | GCTGACAATCATCACCAAGT | GGTTCTCAGCACTATGGGACA |
| ATR | CGCTGAACTGTACGTGGAAA | CAATTAGTGCCTGGTGAACATC |
| BRCA1 | CTGCTCAGGGCTATCCTCTCA | GCTTCTAGTTCAGCCATTTCCTG |
| CCNK | AAAGCCATGTTGGTACTGGGA | TGTAGCCCCAAACGTGTGC |
| E2F1 | CATCAGTACCTGGCCGAGAG | CCCGGGGATTTCACACCTTT |
| FANCD2 | CCCAGAACTGATCAACTCTCCT | CCATCATCACACGGAAGAAA |
| FANCI | CACCACACTTACAGCCCTTG | ATTCCTCCGGAGCTCTGAC |
| RAD51 | GCTGATGAGTTTGGTGTAGCAG | GGAAGACAGGGAGAGTCGTAGA |
| SMARCC2 | GAGGGATAAGCAGGTTCTTCTG | GGGATCCACGTGTCGTAACT |
| TP53 | CCCAAGCAATGGATGATTTGA | GCATTCTGGGAGCTTCATCT |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA |
| dGAPDH | CGTTCATGCCACCACCGCTA | CCACGTCCATCACGCCACAA |
表2 RT-qPCR引物序列
Tab 2 Primer sequence for RT-qPCR
| Primer | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| ATM | GCTGACAATCATCACCAAGT | GGTTCTCAGCACTATGGGACA |
| ATR | CGCTGAACTGTACGTGGAAA | CAATTAGTGCCTGGTGAACATC |
| BRCA1 | CTGCTCAGGGCTATCCTCTCA | GCTTCTAGTTCAGCCATTTCCTG |
| CCNK | AAAGCCATGTTGGTACTGGGA | TGTAGCCCCAAACGTGTGC |
| E2F1 | CATCAGTACCTGGCCGAGAG | CCCGGGGATTTCACACCTTT |
| FANCD2 | CCCAGAACTGATCAACTCTCCT | CCATCATCACACGGAAGAAA |
| FANCI | CACCACACTTACAGCCCTTG | ATTCCTCCGGAGCTCTGAC |
| RAD51 | GCTGATGAGTTTGGTGTAGCAG | GGAAGACAGGGAGAGTCGTAGA |
| SMARCC2 | GAGGGATAAGCAGGTTCTTCTG | GGGATCCACGTGTCGTAACT |
| TP53 | CCCAAGCAATGGATGATTTGA | GCATTCTGGGAGCTTCATCT |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA |
| dGAPDH | CGTTCATGCCACCACCGCTA | CCACGTCCATCACGCCACAA |
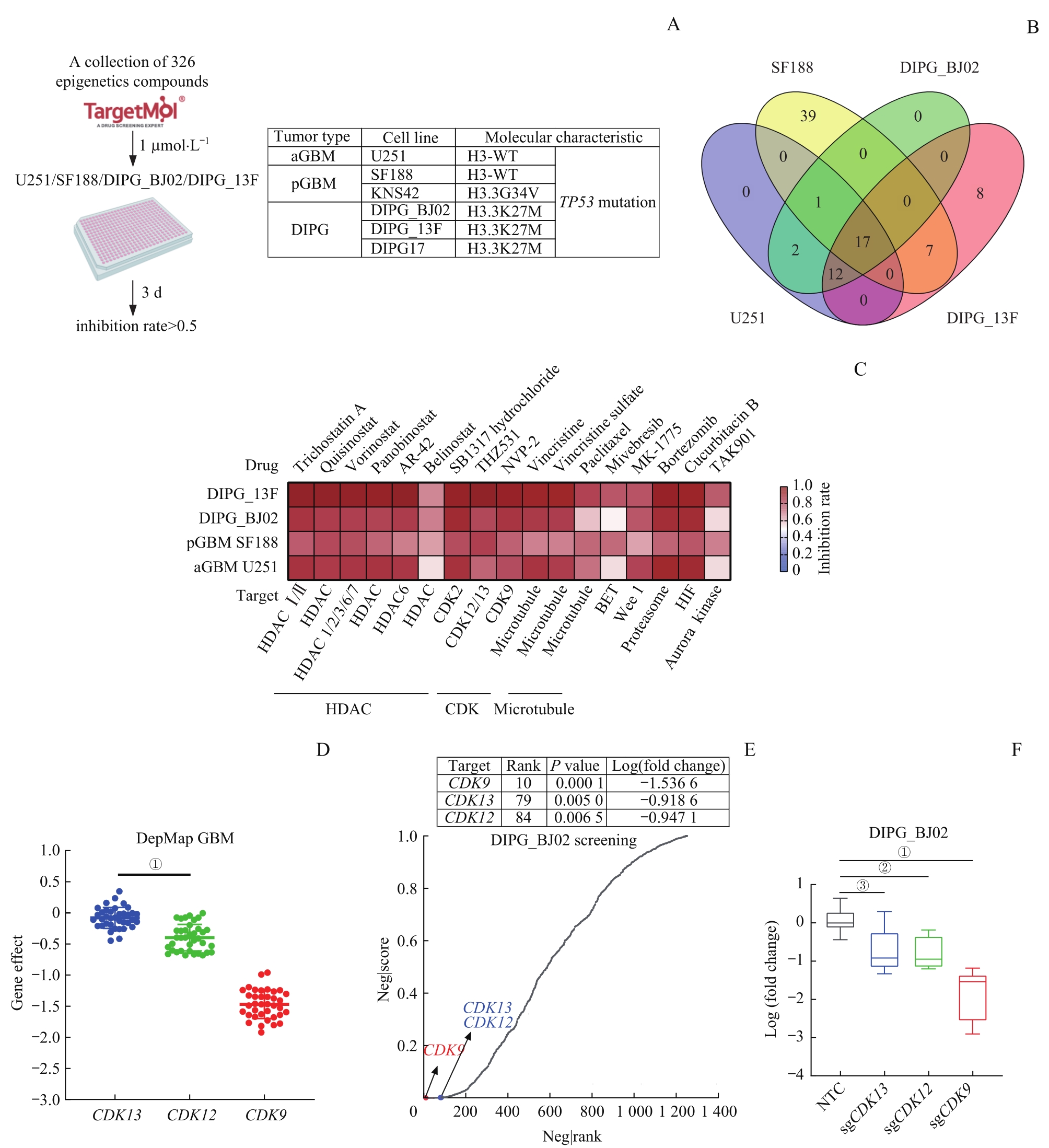
图1 基于表观转录靶向小分子药物库与功能基因组筛选GBM和DIPG共同的潜在药物靶标—— CDK12/13Note: A. Workflow of epigenetic transcription-related targeted small molecule drug library screening in aGBM (U251), pGBM (SF188) and DIPG (DIPG_13F, DIPG_BJ02) cell lines. B. Venn diagram of the epigenetic transcription-related targeted small molecule drug library screening in U251, SF188, DIPG_13F, and DIPG_BJ02 cell lines. C. Heatmap was used to display the screening effects of 17 small molecule classifications. D. Scatter plots showing the dependence of GBM cells on CDK12, CDK13 and CDK9 in DepMap database. E. Epigenetic transcription-related functional genome screening based on CRISPR-Cas9 in vitro for four weeks in DIPG_BJ02 cell line, with the sequencing results analyzed by MAGeCK algorithm. The top 200 genes are thought to be growth-dependent genes in DIPG_BJ02 cell line. F. Box plot of log (fold change) in sgCDK12, sgCDK13 and sgCDK9. NTC—negative control. ①P<0.000 1, ②P=0.001 8, ③P=0.002 9.
Fig 1 Screening for common potential drug targets for GBM and DIPG based on epigenetic transcription-related targeted small molecule drug library and functional genome: CDK12 and CDK13
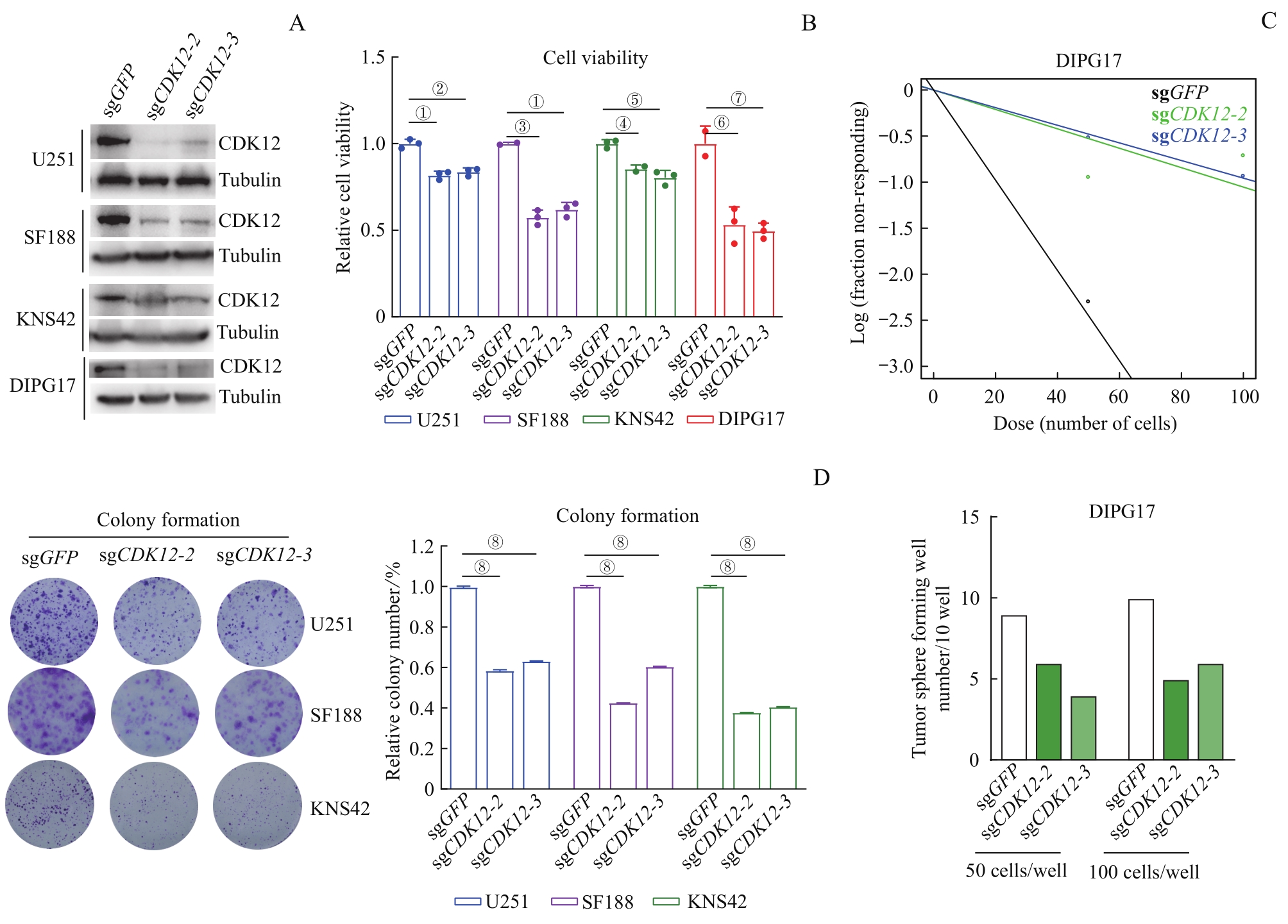
图2 CDK12 敲除对DIPG和GBM肿瘤细胞体外生长的抑制作用Note: A. Western blotting analysis of CDK12 expression levels in aGBM (U251), pGBM (SF188, KNS42), and DIPG17 cell lines after CDK12 knockout by using CRISPR-Cas9 system. B. Detection of cell viability of U251, SF188 and KNS42 on the fourth day, and DIPG17 on the seventh day after CDK12 knockout by CellTiter-Glo. C. Dot plot (upper) and bar chart (below) of neurosphere assay of DIPG17 cells. Fifty or 100 DIPG17 cells were seeded in each well (96-well plate) following CDK12 knockout. After 14 d of culture, the number of neurosphere was counted and mapped. D. Scanogram (left) and cartogram (right) of colony formation assay of CDK12-knockout GBM cell lines (U251, SF188 and KNS42). Cells were plated at 2 000 per well (six well plate), with half-volume mediumeing changed every 4 d, followed by crystal violet staining, photographing and counting in two weeks. ①P=0.000 2, ②P=0.000 4, ③P=0.000 1, ④P=0.008 7, ⑤P=0.001 4, ⑥P=0.004 0, ⑦P=0.002 9,⑧P<0.000 1.
Fig 2 Inhibitory effect of CDK12 knockout on the growth of DIPG and GBM tumor cells in vitro
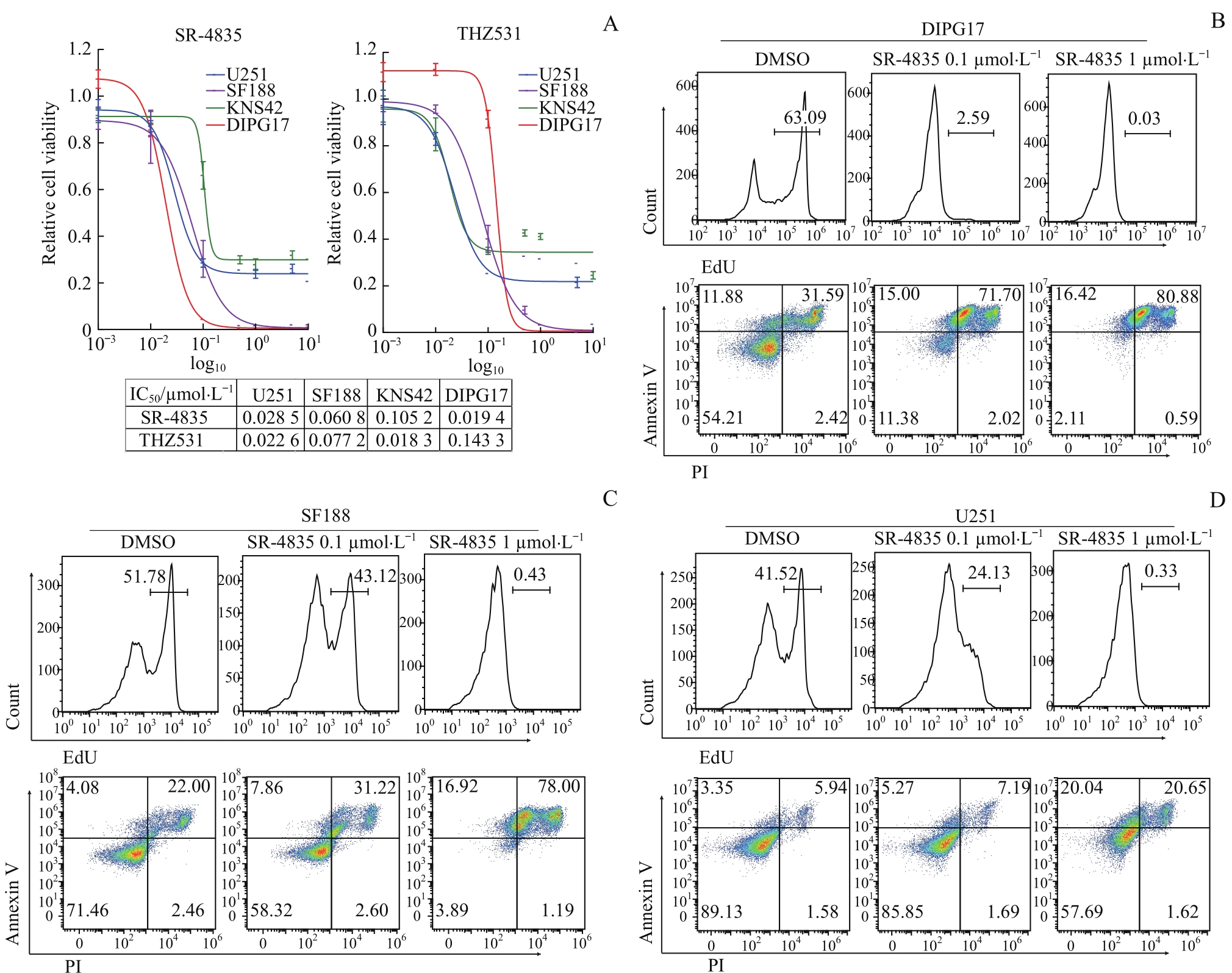
图3 CDK12/13抑制剂对DIPG和GBM体外生长的抑制作用Note: A. Dosage-dependent cell viability curves of CDK12/13 inhibitor SR-4835 or THZ531-treated GBM or DIPG cell lines. B. Detection of proliferation and apoptosis of DIPG17 cells treated with DMSO or SR-4835 (0.1, 1 μmol/L) by flow cytometry. C. Detection of proliferation and apoptosis of SF188 cells treated with DMSO or SR-4835 (0.1, 1 μmol/L) by flow cytometry. D. Detection of proliferation and apoptosis of U251 cells treated with DMSO or SR-4835 (0.1, 1 μmol/L) by flow cytometry. Cell proliferation was detected by EdU-flow cytometry on 24 h and apoptosis was detected by Annexin V/PI-flow cytometry on 48 h.
Fig 3 Inhibitory effect of CDK12/13 inhibitors on the growth of DIPG and GBM in vitro
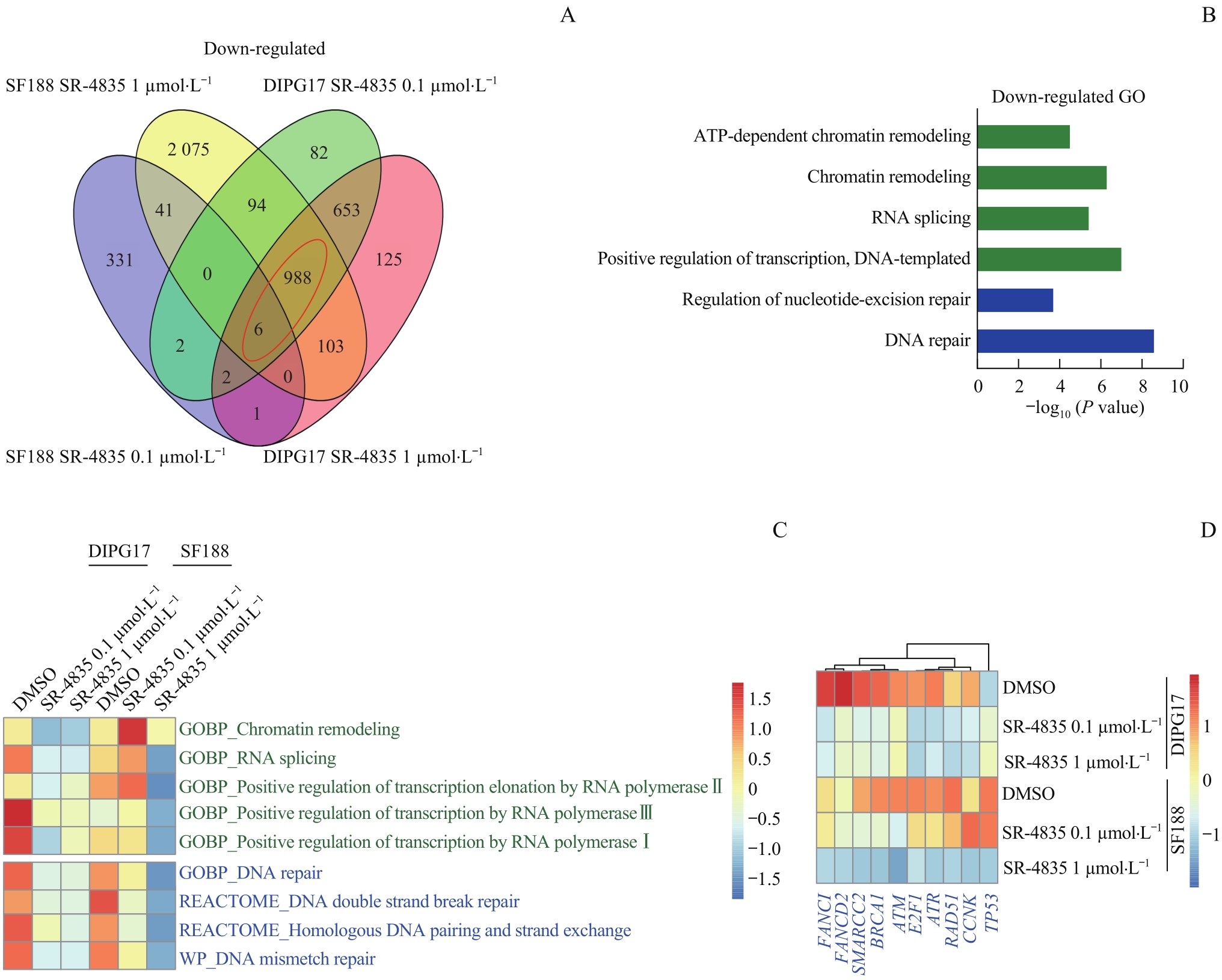
图4 靶向抑制CDK12/13显著降低GBM和DIPG细胞DDR相关基因的富集Note: A. Venn diagram showing differentially expressed genes (Gfold<-1) of 0.1 μmol/L or 1 μmol/L SR-4835 versus DMSO-treated DIPG17 and SF188 cell lines. B. GO analysis on enriched function of down-regulated genes in DIPG17 and SF188 cell lines treated with 0.1 μmol/L or 1 μmol/L SR-4835. C. GSVA analysis of enriched gene sets in DIPG17 and SF188 cell lines. D. Heatmap analysis of RNA-seq data showing transcript level expression alteration of representative DDR-related genes after SR-4835 treatment.
Fig 4 Targeted inhibition of CDK12/13 significantly reduced the enrichment of gene sets involved in DDR in GBM and DIPG cells
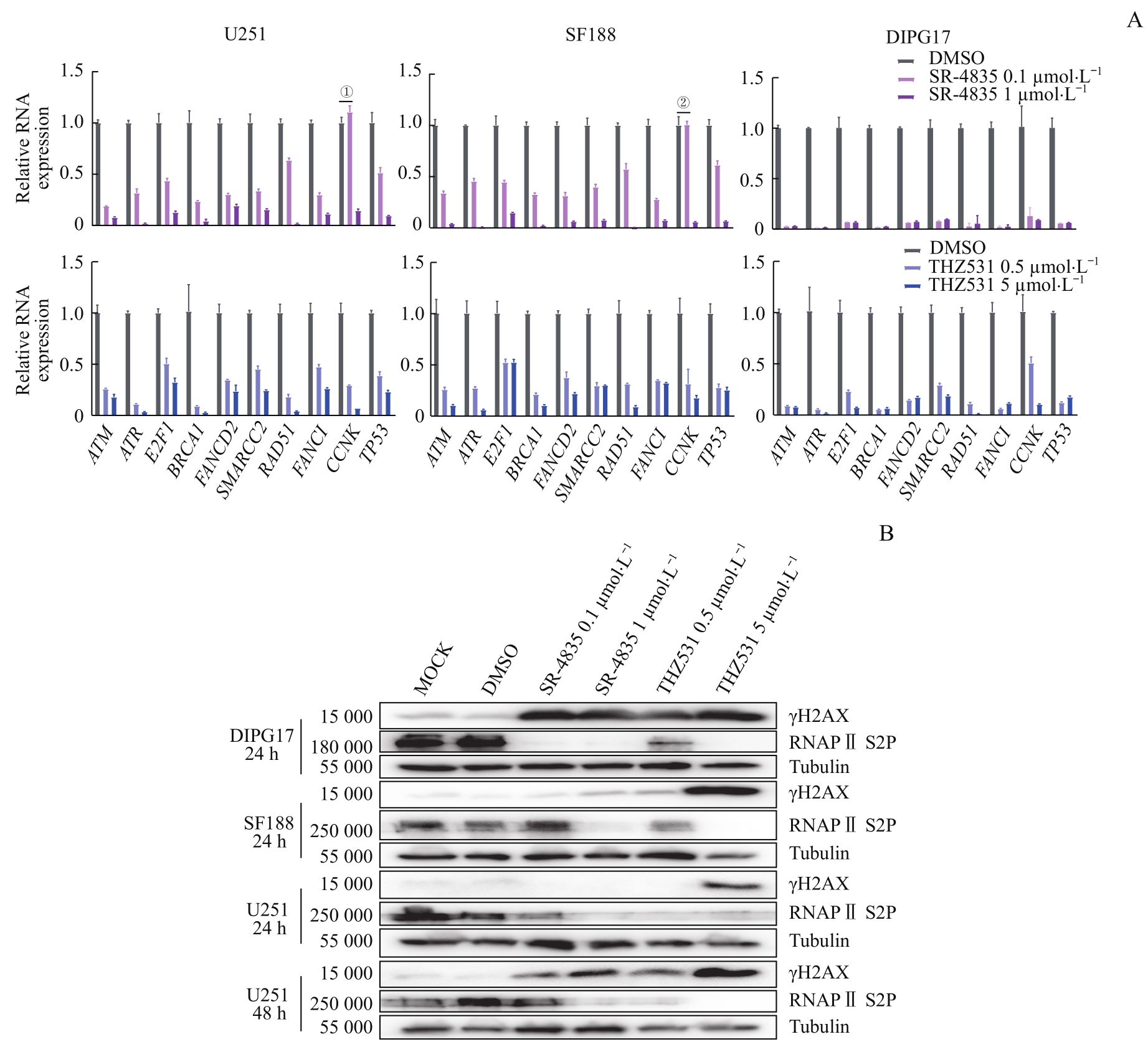
图5 靶向抑制CDK12/13通过显著下调GBM和DIPG细胞DDR相关基因的转录引起DNA损伤的积累Note: A. RT-qPCR results showed that SR-4835 or THZ531 treatment for 20 h had effects on the expression of genes related to DNA damage repair in DIPG17, SF188, and U251 cell lines. B. Western blotting results showed the effect of SR-4835 and THZ531 treatment on protein levels of γH2AX and RNA Pol Ⅱ S2P in DIPG17, SF188, and U251 cell lines. ①P=0.001 8, 0.1 μmol/L SR4835 versus DMSO; ②P>0.05, 0.1 μmol/L SR4835 versus DMSO. Except for the P values shown in ① and ②, all other P values are less than 0.000 1 (0.1 μmol/L or 1 μmol/L SR4835 versus DMSO, 0.5 μmol/L or 5 μmol/L THZ531 versus DMSO).
Fig 5 Targeted inhibition of CDK12/13 induced accumulation of DNA damage by significantly down-regulating transcription of genes involved in DDR in GBM and DIPG cells
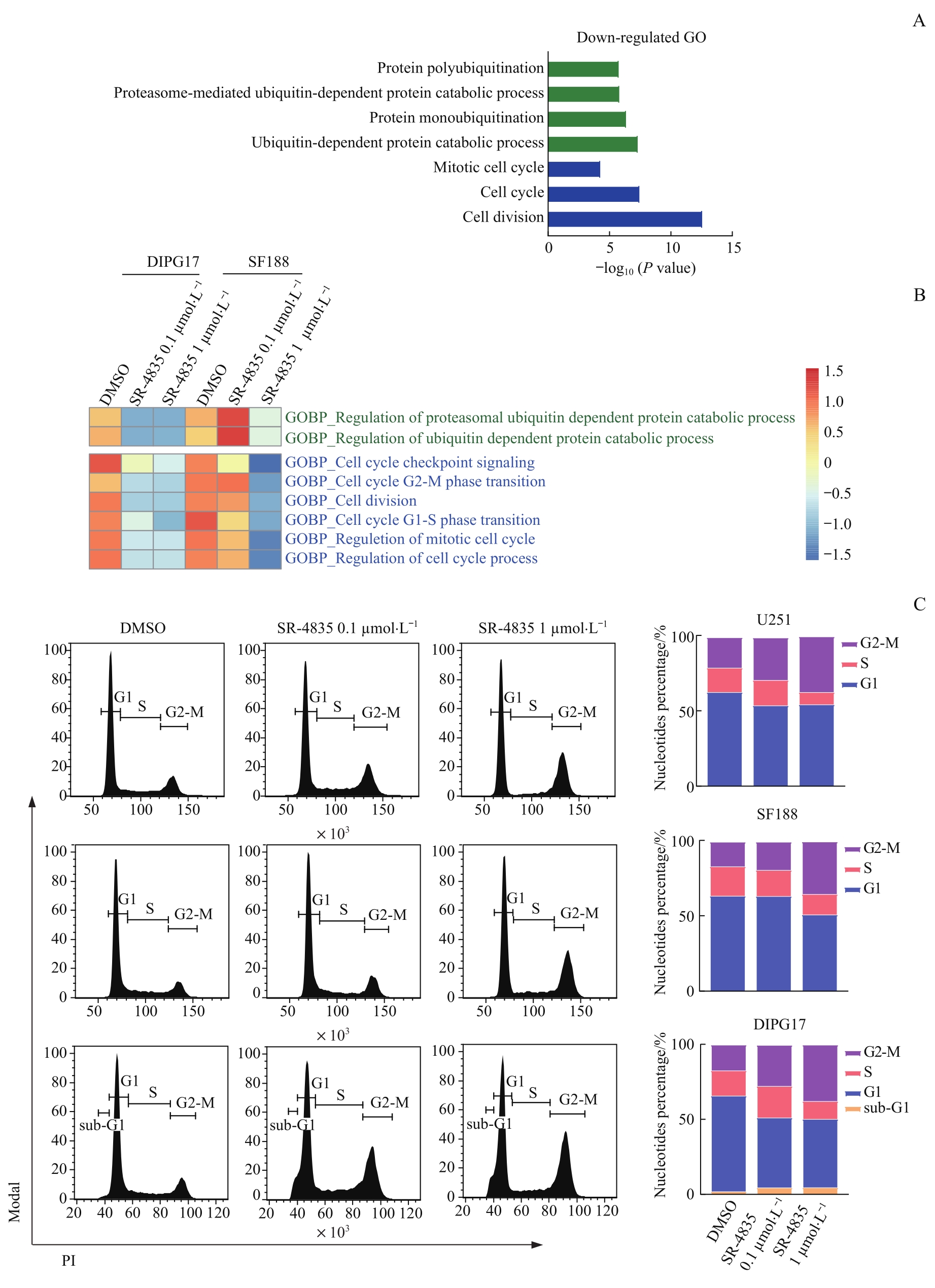
图6 靶向抑制CDK12/13诱导DIPG和GBM细胞发生G2-M细胞周期阻滞Note: A. GO analysis on enriched function of down-regulated genes in DIPG17 and SF188 cells. B. Heatmap showing GSVA analysis of enriched ubiquitin and cell cycle-related gene sets in DIPG17 and SF188 cells. C. Flow cytometry and bar charts analyses on cell cycle of U251, SF188, and DIPG17 cells treated with DMSO, 0.1 μmol/L SR-4835 or 1 μmol/L SR-4835 for 24 h.
Fig 6 Targeted inhibition of CDK12/13 induced G2-M cell cycle arrest in DIPG and GBM cells
| 1 | MILLER K D, OSTROM Q T, KRUCHKO C, et al. Brain and other central nervous system tumor statistics, 2021[J]. CA Cancer J Clin, 2021, 71(5): 381-406. |
| 2 | CHOI S, YU Y, GRIMMER M R, et al. Temozolomide-associated hypermutation in gliomas[J]. Neuro Oncol, 2018, 20(10): 1300-1309. |
| 3 | OSTROM Q T, PRICE M, NEFF C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015—2019[J]. Neuro Oncol, 2022, 24(Suppl 5): v1-v95. |
| 4 | MCKINNON C, NANDHABALAN M, MURRAY S A, et al. Glioblastoma: clinical presentation, diagnosis, and management[J]. BMJ, 2021, 374: n1560. |
| 5 | BRENNAN C W, VERHAAK R G, MCKENNA A, et al. The somatic genomic landscape of glioblastoma[J]. Cell, 2013, 155(2): 462-477. |
| 6 | MENG W, WANG J J, WANG B C, et al. CDK7 inhibition is a novel therapeutic strategy against GBM both in vitro and in vivo[J]. Cancer Manag Res, 2018, 10: 5747-5758. |
| 7 | YAN G, WANG Y F, CHEN J C, et al. Advances in drug development for targeted therapies for glioblastoma[J]. Med Res Rev, 2020, 40(5): 1950-1972. |
| 8 | NJONKOU R, JACKSON C M, WOODWORTH G F, et al. Pediatric glioblastoma: mechanisms of immune evasion and potential therapeutic opportunities[J]. Cancer Immunol Immunother, 2022, 71(8): 1813-1822. |
| 9 | BERGER T R, WEN P Y, LANG-ORSINI M, et al. World Health Organization 2021 classification of central nervous system tumors and implications for therapy for adult-type gliomas: a review[J]. JAMA Oncol, 2022, 8(10): 1493-1501. |
| 10 | ŚLEDZIŃSKA P, BEBYN M G, FURTAK J, et al. Prognostic and predictive biomarkers in gliomas[J]. Int J Mol Sci, 2021, 22(19): 10373. |
| 11 | PATHANIA M, DE JAY N, MAESTRO N, et al. H3.3K27M cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas[J]. Cancer Cell, 2017, 32(5): 684-700.e9. |
| 12 | HOFFMAN L M, VELDHUIJZEN VAN ZANTEN S E M, COLDITZ N, et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): a collaborative report from the international and European society for pediatric oncology DIPG registries[J]. J Clin Oncol, 2018, 36(19): 1963-1972. |
| 13 | AZIZ-BOSE R, MONJE M. Diffuse intrinsic pontine glioma: molecular landscape and emerging therapeutic targets[J]. Curr Opin Oncol, 2019, 31(6): 522-530. |
| 14 | RASHED W M, MAHER E, ADEL M, et al. Pediatric diffuse intrinsic pontine glioma: where do we stand?[J]. Cancer Metastasis Rev, 2019, 38(4): 759-770. |
| 15 | COONEY T M, LUBANSZKY E, PRASAD R, et al. Diffuse midline glioma: review of epigenetics[J]. J Neurooncol, 2020, 150(1): 27-34. |
| 16 | MENG W, WANG B C, MAO W W, et al. Enhanced efficacy of histone deacetylase inhibitor combined with bromodomain inhibitor in glioblastoma[J]. J Exp Clin Cancer Res, 2018, 37(1): 241. |
| 17 | MO J L, TAN K Z, DONG Y, et al. Therapeutic targeting the oncogenic driver EWSR1-FLI1 in Ewing sarcoma through inhibition of the FACT complex[J]. Oncogene, 2023, 42(1): 11-25. |
| 18 | NAGARAJA S, VITANZA N A, WOO P J, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma[J]. Cancer Cell, 2017, 31(5): 635-652.e6. |
| 19 | NGUYEN T T T, ZHANG Y R, SHANG E Y, et al. HDAC inhibitors elicit metabolic reprogramming by targeting super-enhancers in glioblastoma models[J]. J Clin Invest, 2020, 130(7): 3699-3716. |
| 20 | ANASTAS J N, ZEE B M, KALIN J H, et al. Re-programing chromatin with a bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG[J]. Cancer Cell, 2019, 36(5): 528-544.e10. |
| 21 | DAHL N A, DANIS E, BALAKRISHNAN I, et al. Super elongation complex as a targetable dependency in diffuse midline glioma[J]. Cell Rep, 2020, 31(1): 107485. |
| 22 | RANJAN A, PANG Y, BUTLER M, et al. Targeting CDK9 for the treatment of glioblastoma[J]. Cancers (Basel), 2021, 13(12): 3039. |
| 23 | CHOU J, QUIGLEY D A, ROBINSON T M, et al. Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy[J]. Cancer Discov, 2020, 10(3): 351-370. |
| 24 | LIANG K W, GAO X, GILMORE J M, et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing[J]. Mol Cell Biol, 2015, 35(6): 928-938. |
| 25 | CHOI S H, KIM S, JONES K A. Gene expression regulation by CDK12: a versatile kinase in cancer with functions beyond CTD phosphorylation[J]. Exp Mol Med, 2020, 52(5): 762-771. |
| 26 | QUEREDA V, BAYLE S, VENA F, et al. Therapeutic targeting of CDK12/CDK13 in triple-negative breast cancer[J]. Cancer Cell, 2019, 36(5): 545-558.e7. |
| 27 | WANG C, WANG H, LIEFTINK C, et al. CDK12 inhibition mediates DNA damage and is synergistic with sorafenib treatment in hepatocellular carcinoma[J]. Gut, 2020, 69(4): 727-736. |
| 28 | DIETER S M, SIEGL C, CODÓ P L, et al. Degradation of CCNK/CDK12 is a druggable vulnerability of colorectal cancer[J]. Cell Rep, 2021, 36(3): 109394. |
| 29 | LIU H, SHIN S H, CHEN H Y, et al. CDK12 and PAK2 as novel therapeutic targets for human gastric cancer[J]. Theranostics, 2020, 10(14): 6201-6215. |
| 30 | JIANG B S, JIANG J, KALTHEUNER I H, et al. Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma[J]. Eur J Med Chem, 2021, 221: 113481. |
| 31 | BLAZEK D, KOHOUTEK J, BARTHOLOMEEUSEN K, et al. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes[J]. Genes Dev, 2011, 25(20): 2158-2172. |
| 32 | DUBBURY S J, BOUTZ P L, SHARP P A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation[J]. Nature, 2018, 564(7734): 141-145. |
| 33 | ZHANG T H, KWIATKOWSKI N, OLSON C M, et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors[J]. Nat Chem Biol, 2016, 12(10): 876-884. |
| 34 | FENG J X, MEYER C A, WANG Q, et al. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data[J]. Bioinformatics, 2012, 28(21): 2782-2788. |
| 35 | INIGUEZ A B, STOLTE B, WANG E J, et al. EWS/FLI confers tumor cell synthetic lethality to CDK12 inhibition in Ewing sarcoma[J]. Cancer Cell, 2018, 33(2): 202-216.e6. |
| 36 | COLLINS P L, PURMAN C, PORTER S I, et al. DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner[J]. Nat Commun, 2020, 11(1): 3158. |
| 37 | LI R, OKADA H, YAMASHITA T, et al. FOXM1 is a novel molecular target of AFP-positive hepatocellular carcinoma abrogated by proteasome inhibition[J]. Int J Mol Sci, 2022, 23(15): 8305. |
| 38 | HOPKINS J L, ZOU L. Induction of BRCAness in triple-negative breast cancer by a CDK12/13 inhibitor improves chemotherapy[J]. Cancer Cell, 2019, 36(5): 461-463. |
| 39 | NIU T, LI K L, JIANG L, et al. Noncovalent CDK12/13 dual inhibitors-based PROTACs degrade CDK12-cyclin K complex and induce synthetic lethality with PARP inhibitor[J]. Eur J Med Chem, 2022, 228: 114012. |
| 40 | YAM C Q X, LIM H H, SURANA U. DNA damage checkpoint execution and the rules of its disengagement[J]. Front Cell Dev Biol, 2022, 10: 1020643. |
| 41 | XIE J B, SHEN Z Y, ANRAKU Y, et al. Nanomaterial-based blood-brain-barrier (BBB) crossing strategies[J]. Biomaterials, 2019, 224: 119491. |
| 42 | BOBO R H, LASKE D W, AKBASAK A, et al. Convection-enhanced delivery of macromolecules in the brain[J]. Proc Natl Acad Sci U S A, 1994, 91(6): 2076-2080. |
| 43 | MADANI F, LINDBERG S, LANGEL U, et al. Mechanisms of cellular uptake of cell-penetrating peptides[J]. J Biophys, 2011, 2011: 414729. |
| 44 | HERAVI SHARGH V, LUCKETT J, BOUZINAB K, et al. Chemosensitization of temozolomide-resistant pediatric diffuse midline glioma using potent nanoencapsulated forms of a N(3)-propargyl analogue[J]. ACS Appl Mater Interfaces, 2021, 13(30): 35266-35280. |
| 45 | WU T T, LIU Y, CAO Y, et al. Engineering macrophage exosome disguised biodegradable nanoplatform for enhanced sonodynamic therapy of glioblastoma[J]. Adv Mater, 2022, 34(15): e2110364. |
| [1] | 那迪娜·帕尔哈提null, 严妍, 车千纪, 罗菁, 刘鑫男, 李斌. 嵌合抗原受体T细胞疗法在胶质母细胞瘤中的应用与展望[J]. 上海交通大学学报(医学版), 2021, 41(7): 982-986. |
| [2] | 刘景景, 胡慧洁, 刘子楷, 宋明柯. 钠钙交换体阻滞剂CB-DMB对人胶质母细胞瘤细胞生长的抑制作用[J]. 上海交通大学学报(医学版), 2021, 41(6): 710-716. |
| [3] | 李 超,糜坚青,王 瑾. 费城染色体样急性淋巴细胞白血病的研究进展[J]. 上海交通大学学报(医学版), 2020, 40(9): 1294-1301. |
| [4] | 张辉林,朱晓娜,杨烁,刘朦迪,朱迪,余韵. Fbxo22基因敲除小鼠模型的建立和表型研究[J]. 上海交通大学学报(医学版), 2019, 39(4): 353-. |
| [5] | 翁震 1,钮晓音 2,周俊松 1. CRISPR-Cas9技术在非肿瘤性血液病中的应用[J]. 上海交通大学学报(医学版), 2018, 38(11): 1396-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||