
上海交通大学学报(医学版) ›› 2023, Vol. 43 ›› Issue (9): 1115-1130.doi: 10.3969/j.issn.1674-8115.2023.09.006
收稿日期:2023-01-29
接受日期:2023-08-03
出版日期:2023-09-28
发布日期:2023-09-28
通讯作者:
张 磊,电子信箱:weiymzhl@126.com。作者简介:刘思雨(1998—),女,硕士生;电子信箱:lsy19982021@163.com。
基金资助:Received:2023-01-29
Accepted:2023-08-03
Online:2023-09-28
Published:2023-09-28
Contact:
ZHANG Lei, E-mail: weiymzhl@126.com.Supported by:摘要:
目的·基于单细胞转录组测序(single-cell RNA sequencing,scRNA-seq)探究单次和重复七氟烷麻醉对新生小鼠前额叶皮层(prefrontal cortex,PFC)神经元发育的损伤机制。方法·将新生小鼠分为重复麻醉暴露(Sev3)组、单次麻醉暴露(Sev1)组和对照组,每组3只。Sev3组于出生后第6、第7、第8日接受3%七氟烷和60%氧气的麻醉,Sev1组仅在出生后第6日接受麻醉,出生后第9日获取3组小鼠的PFC进行scRNA-seq。通过UMAP(uniform manifold approximation and projection)聚类、RNA速度分析、转录因子分析(SCENIC)获得七氟烷麻醉后新生小鼠PFC细胞图谱和神经元细分亚群图谱,并进行差异表达基因分析;通过基因本体论(Gene Ontology,GO)和京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)数据库分析研究差异基因富集的生物学过程和通路;利用QuSAGE分析描述细胞周期及Hippo信号通路基因集的激活情况。通过CytoTRACE评分判断七氟烷麻醉后新生小鼠PFC神经元谱系细胞转录本丰富度和干性;利用拟时序分析(pseudo-time analysis)确定神经元分化轨迹,通过分支表达分析模型(BEAM)对发育节点进行解析,以确定决定细胞不同命运的关键基因。结果·3组新生小鼠PFC经scRNA-seq共获得40 061个细胞,10种细胞类型。单次七氟烷麻醉后PFC细胞下调的基因富集在细胞分化、前脑神经元分化、去甲肾上腺素能神经元分化和大脑皮层γ-氨基丁酸能中间神经元分化。重复七氟烷麻醉后下调基因富集在细胞分化的正向调节。单次七氟烷麻醉后下调的差异基因富集的KEGG通路有转化生长因子β信号通路。重复七氟烷麻醉后下调的差异基因富集的KEGG通路有Notch信号通路。SCENIC显示,早期生长反应因子1(early growth response 1,Egr1)、SRY-box转录因子7(SRY-box transcription factor 7,Sox7)在单次和重复七氟烷麻醉后均上调(均P<0.01),HES家族bHLH转录因子6(HES family bHLH transcription factor 6,Hes6)、NKX同源框-1基因(NK2 homeobox 1,Nkx2-1)仅在单次七氟烷麻醉后下调(均P<0.01)。七氟烷麻醉后放射状胶质细胞和神经元与细胞周期相关的基因集激活度增加,并且重复麻醉暴露后激活度增加更明显。重复七氟烷麻醉后神经元与Hippo信号通路相关的基因集由抑制变为激活。将8 224个神经元单独进行亚群分析获得8种神经元谱系细胞,CytoTRACE评分表明七氟烷麻醉后神经元干性增加,神经元发育迟滞。PFC神经元经拟时序分析区分为3个发育阶段,重复七氟烷麻醉使PFC神经元分化拟时间后退(P=0.000)。单次七氟烷麻醉暴露后新生小鼠PFC神经元下调的基因富集在细胞周期蛋白依赖性蛋白丝氨酸/苏氨酸激酶活性的调节、有丝分裂细胞周期相变、细胞分化、长期记忆、有丝分裂细胞周期G1/S转换。重复七氟烷麻醉暴露后PFC神经元下调基因富集在细胞群增殖的负向调控、细胞分化的正向调节、前脑神经元的分化、典型Wnt信号通路的正向调控和细胞分化。结论·单次和重复七氟烷麻醉均促进新生小鼠PFC神经祖细胞增殖和神经元迁移,重复七氟烷麻醉抑制PFC的神经祖细胞向神经元分化,其潜在机制可能与细胞周期转换有关。
中图分类号:
刘思雨, 张磊. 七氟烷抑制新生小鼠前额叶皮质神经祖细胞向神经元分化发育[J]. 上海交通大学学报(医学版), 2023, 43(9): 1115-1130.
LIU Siyu, ZHANG Lei. Sevoflurane inhibits the differentiation and development of neural progenitor cells into neurons in the prefrontal cortex of newborn mice[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(9): 1115-1130.

图1 各组小鼠PFC细胞图谱、细胞类型分布及占比Note: A. Experimental procedures. The PFC cells acquired on postnatal day 9 (P9) from control (Ctrl) group, single anesthesia exposure (Sev1) group, and multiple anesthesia exposure (Sev3) group were prepared in single-cell suspension for 10x Genomics scRNA-seq. B. UMAP cluster showing PFC scRNA-seq results. C. Clustering visualization of RNA velocity analysis results of PFC cells. D. Bubble map of sorting gene markers in PFC cells. Bubble size represents the percentage of cells with the gene expressed in the population, and bubble colour represents the average value of the gene expressed in the population. E. Distribution of 3 groups of sample cells on the UMAP plot. F. Bar graphs of the percentages of cell types in 9 samples.
Fig 1 Cellular profiles, and cell type distribution and proportion in the PFCs of three groups of mice

图2 新生小鼠七氟烷麻醉后PFC细胞转录组变化和功能注释分析Note: A. Volcano map showing differentially expressed genes between Sev1 group and Ctrl group. B/C. Top 15 enrichment entries of down-regulated genes (B) and up-regulated genes (C) in the GO-BP database between Sev1 group and Ctrl group. D. Volcano map showing differentially expressed genes between Sev3 group and Ctrl group. E/F. Top 15 enrichment entries of down-regulated genes (E) and up-regulated genes (F) in the GO-BP database between Sev3 group and Ctrl group. Entries highlighted in red represent biological processes associated with PFC neurogenesis.
Fig 2 Analysis of transcriptome changes and functional annotation of PFC cells in neonatal mice after sevoflurane anesthesia
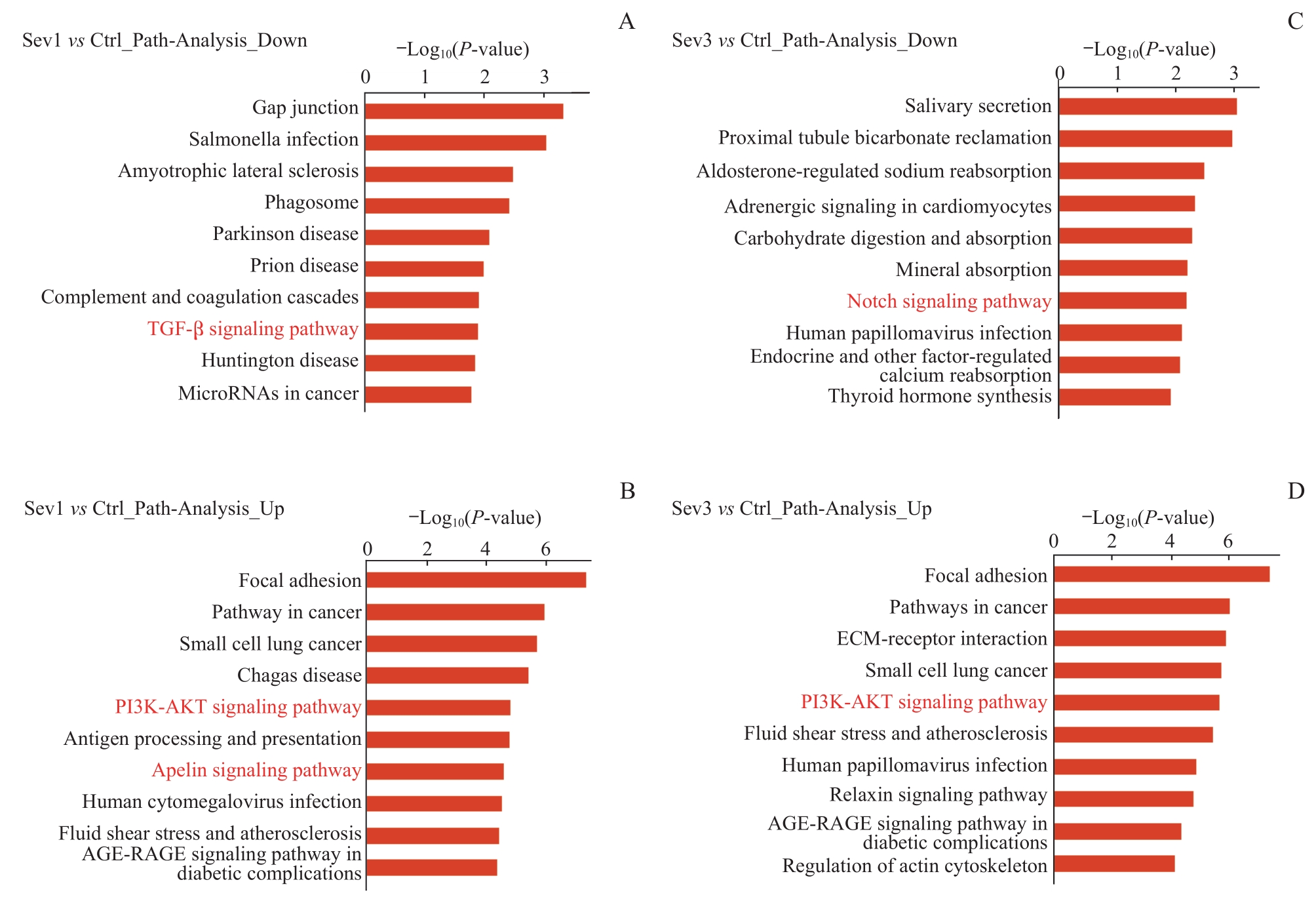
图3 新生小鼠七氟烷麻醉后PFC细胞差异表达基因的通路分析Note: A/B. Top 10 enrichment pathway entries of down-regulated genes (A) and up-regulated genes (B) in the KEGG database between Sev1 group and Ctrl group. C/D. Top 10 enrichment pathway entries of down-regulated genes (C) and up-regulated genes (D) in the KEGG database between Sev3 group and Ctrl group. Entries marked in red represent pathways associated with PFC neurogenesis.
Fig 3 Pathway analysis of differential expressed genes in PFC cells in neonatal mice after sevoflurane anesthesia

图4 新生小鼠七氟烷麻醉后PFC细胞转录因子变化Note: A. AUCell values of Egr1. B. AUCell values of Sox7. C. AUCell values of Hes6. D. AUCell values of Nkx2-1. ①P=0.000, ②P=0.009. ns—not significant.
Fig 4 Transcription factor changes in PFC cells in neonatal mice after sevoflurane anesthesia
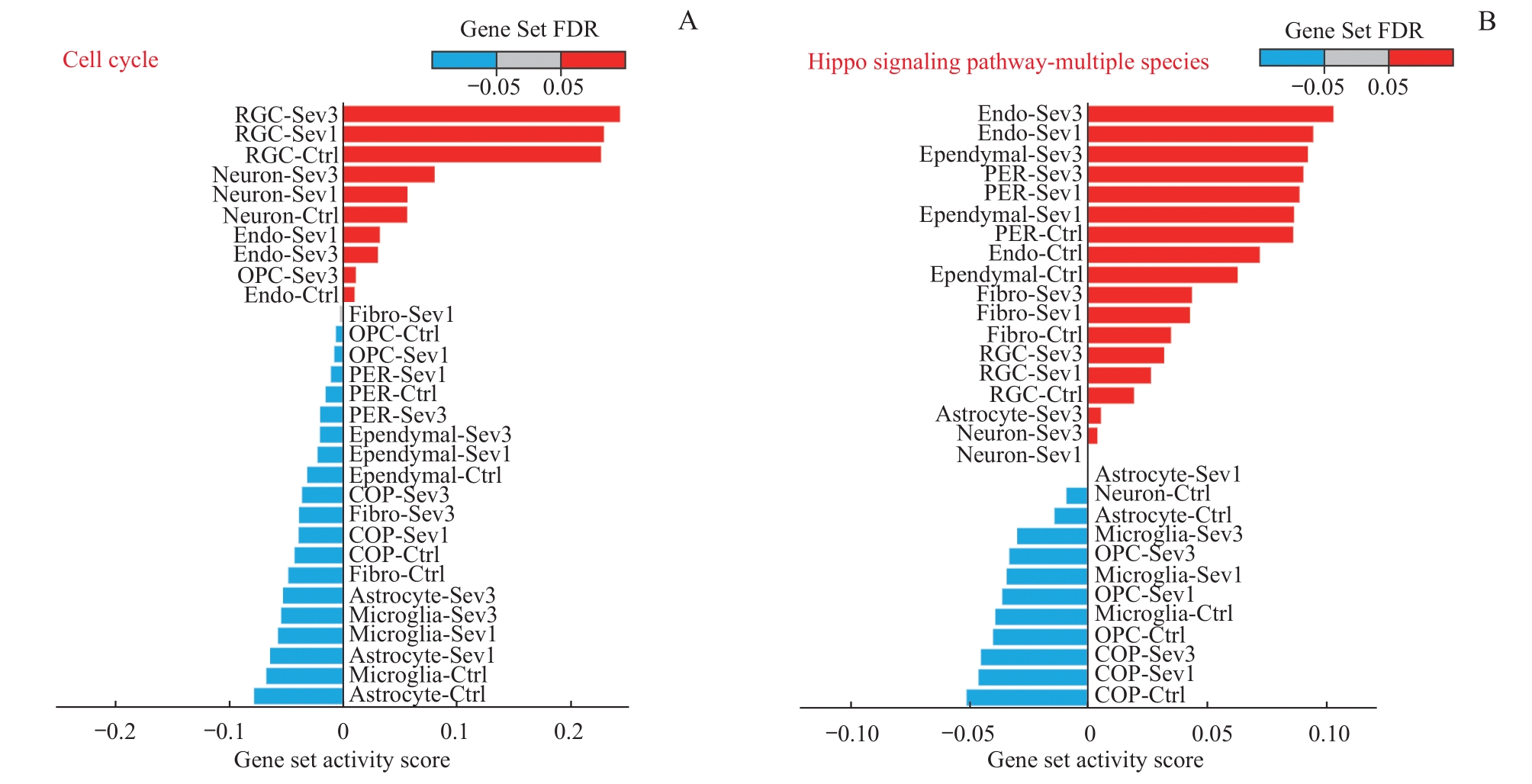
图5 新生小鼠七氟烷麻醉后PFC细胞细胞周期和Hippo信号通路的基因集激活情况Note: QuSAGE analysis of differentially expressed genes after exposure to sevoflurane anesthesia. A. Cell cycle pathway gene set activation analysis. B. Hippo signaling pathway-multiple species gene set activation analysis.
Fig 5 Gene set activation of cell cycle pathways and Hippo signaling pathways in PFC cells in neonatal mice after sevoflurane anesthesia
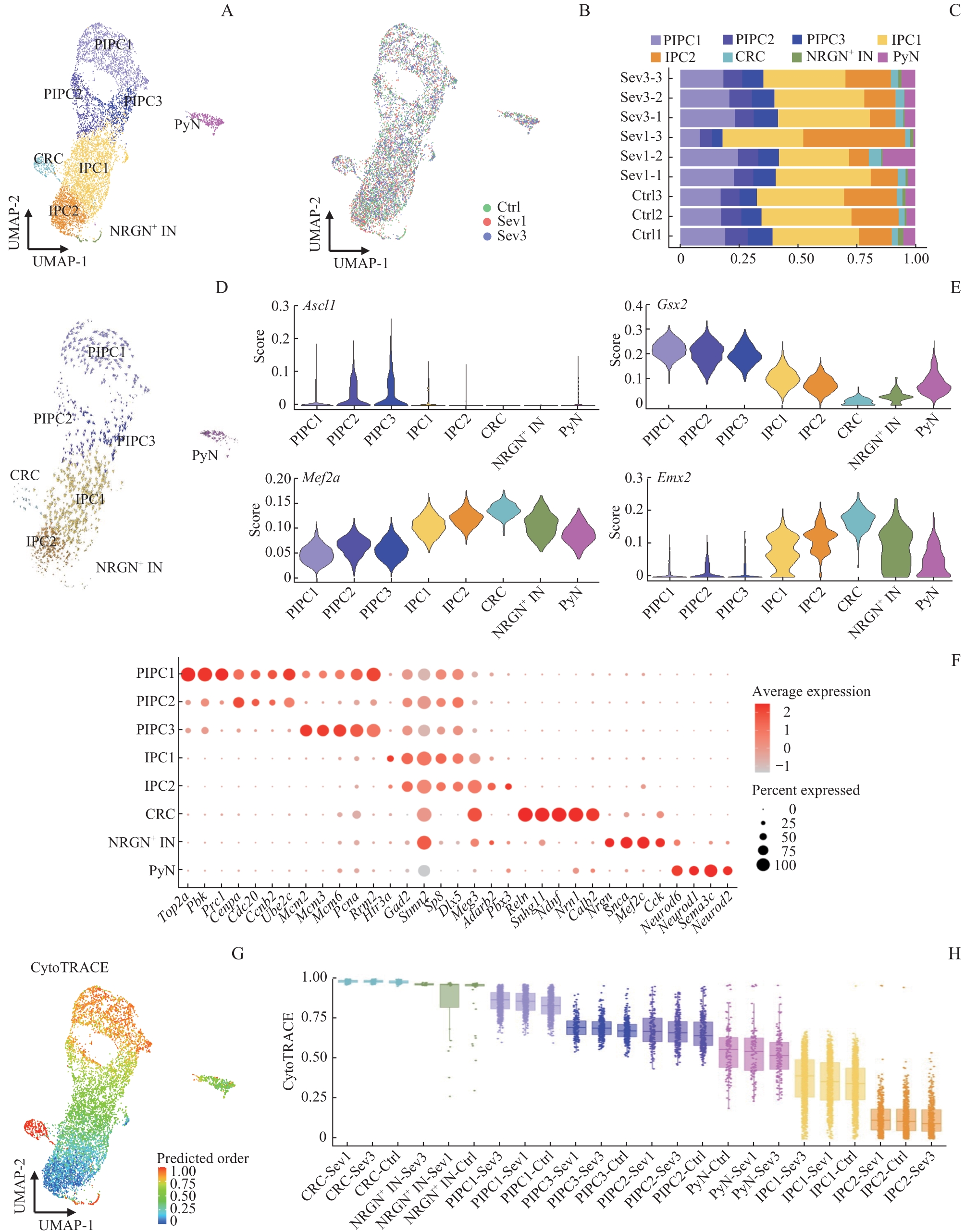
图6 各组小鼠PFC神经元图谱、类型分布及干性分析Note: A. UMAP cluster showing the scRNA-seq results of PFC cells. B. Distribution of 3 groups of sample cells on the UMAP plot. C. Bar graphs of the percentages of neuron subdivisions in 9 samples. D. Clustering visualization of RNA velocity analysis results of PFC neurons. E. SCENIC analysis. F. Bubble map of sorting gene markers in PFC neurons. Bubble size represents the percentage of cells with the gene expressed in the population, and bubble colour represents the average value of the gene expressed in the population. G. CytoTRACE score. The color represents the score, and the higher the score, the higher the cell stemness. H. CytoTRACE box plot.
Fig 6 PFC neuron profiles, type distribution and stemness analysis of mice in 3 groups
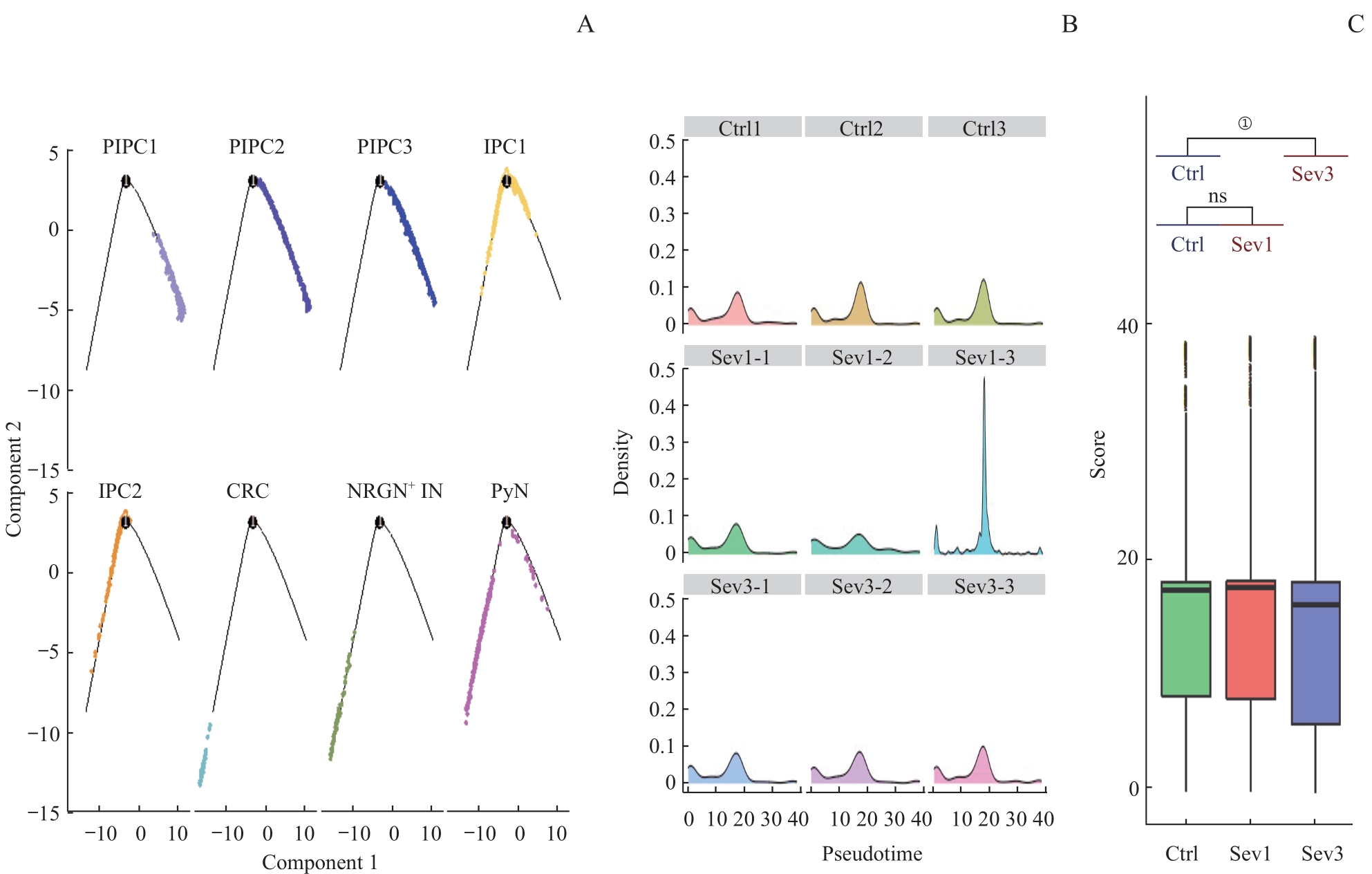
图7 七氟烷麻醉后新生小鼠PFC神经元谱系细胞拟时序分析Note: A. Cell trajectory skeleton plots. B. Density profiles. The horizontal axis represents the proposed time, and the vertical axis represents the density of cell distribution. C. Boxplot of the pseudotime of PFC neuron development in 3 groups. ①P=0.000.
Fig 7 Pseudo‐time analysis of PFC neuronal lineage cells in neonatal mice after sevoflurane anesthesia

图8 七氟烷麻醉后新生小鼠PFC神经元谱系细胞BEAM分析热图Note: Left: heat map of top 1 000 genes that determine cell fate. Middle: top 15 enrichment entries of the 1 000 genes that determine cell fate in the GO-BP database. Right: heat map of Beam analysis of genes with significant differences in PFC neurons after sevoflurane anesthesia. The horizontal axis represents the proposed time, and the vertical axis represents the clustering of genes. The early developmental cluster included PIPC1?3, the mid-developmental cluster included IPC1?2, and the late developmental cluster included CRC, NRGN+ IN, and PyN.
Fig 8 BEAM analysis heat map of PFC neuronal lineage cells in neonatal mice after sevoflurane anesthesia
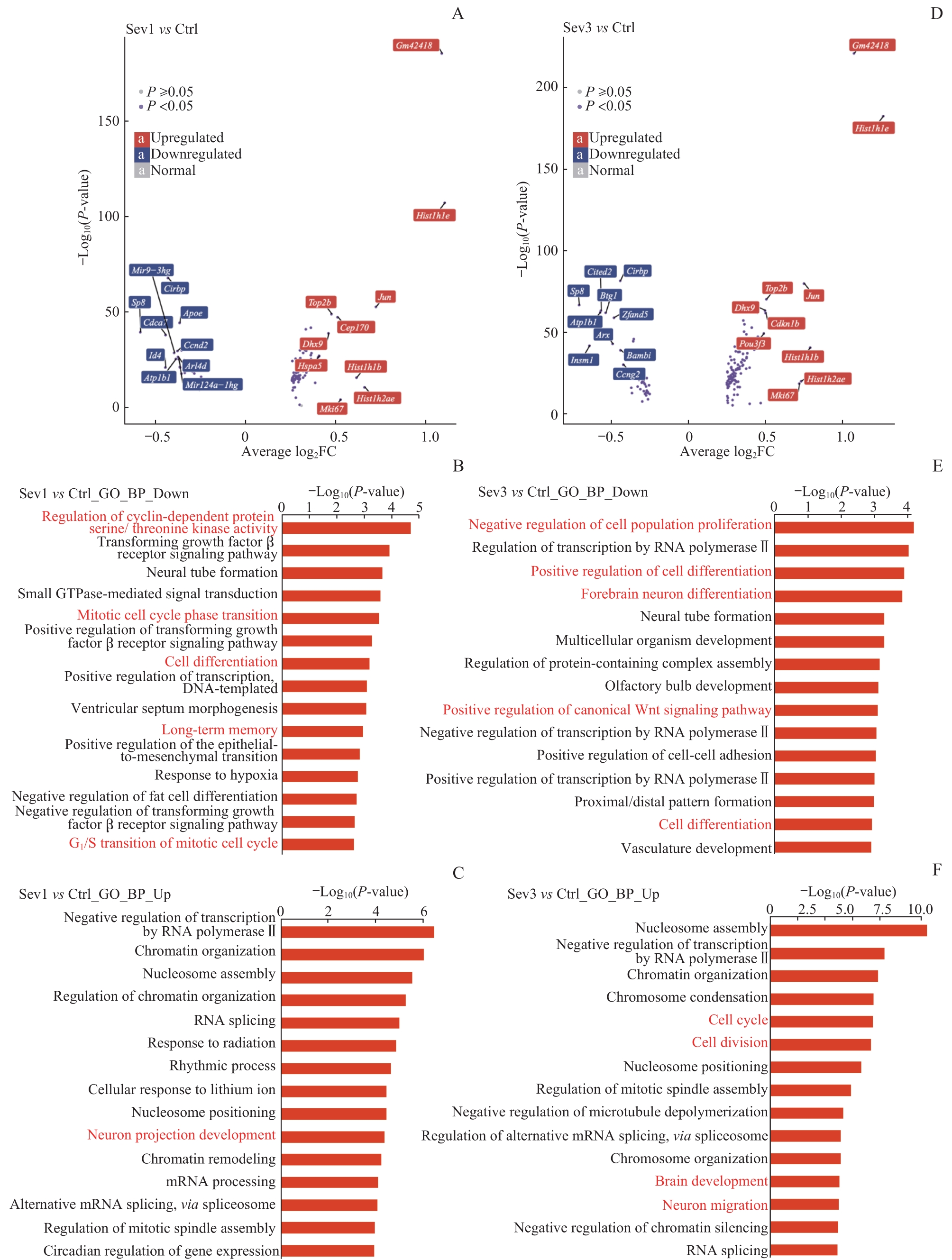
图9 新生小鼠七氟烷麻醉后PFC神经元的转录组变化和功能注释分析Note:A. Volcano map showing differentially expressed genes between Sev1 group and Ctrl group. B/C. Top 15 enrichment entries of down-regulated genes (B) and up-regulated genes (C) in the GO-BP database between Sev1 group and Ctrl group. D. Volcano map showing differentially expressed genes between Sev3 group and Ctrl group. E/F. Top 15 enrichment entries of down-regulated genes (E) and up-regulated genes (F) in the GO-BP database between Sev3 group and Ctrl group. Entries highlighted in red represent biological processes associated with PFC neurogenesis.
Fig 9 Analysis of transcriptome changes and functional annotation of PFC neurons in neonatal mice after sevoflurane anesthesia
| 1 | SHI Y W, WANG G, LI J Y, et al. Hydrogen gas attenuates sevoflurane neurotoxicity through inhibiting nuclear factor κ-light-chain-enhancer of activated B cells signaling and proinflammatory cytokine release in neonatal rats[J]. Neuroreport, 2017, 28(17): 1170-1175. |
| 2 | TIAN Y, CHEN K Y, LIU L D, et al. Sevoflurane exacerbates cognitive impairment induced by Aβ1-40 in rats through initiating neurotoxicity, neuroinflammation, and neuronal apoptosis in rat hippocampus[J]. Mediators Inflamm, 2018, 2018: 3802324. |
| 3 | WU L Z, ZHAO H L, WENG H, et al. Lasting effects of general anesthetics on the brain in the young and elderly: mixed picture of neurotoxicity, neuroprotection and cognitive impairment[J]. J Anesth, 2019, 33(2): 321-335. |
| 4 | ZHAO S, FAN Z Q, HU J, et al. The differential effects of isoflurane and sevoflurane on neonatal mice[J]. Sci Rep, 2020, 10(1): 19345. |
| 5 | OLUTOYE O A, BAKER B W, BELFORT M A, et al. Food and Drug Administration warning on anesthesia and brain development: implications for obstetric and fetal surgery[J]. Am J Obstet Gynecol, 2018, 218(1): 98-102. |
| 6 | CHEN Q C, CHU W, SHENG R, et al. Maternal anesthesia with sevoflurane during the mid-gestation induces social interaction deficits in offspring C57BL/6 mice[J]. Biochem Biophys Res Commun, 2021, 553: 65-71. |
| 7 | FANG F, SONG R X, LING X M, et al. Multiple sevoflurane anesthesia in pregnant mice inhibits neurogenesis of fetal hippocampus via repressing transcription factor PAX6[J]. Life Sci, 2017, 175: 16-22. |
| 8 | LIANG L R, ZENG T, ZHAO Y Y, et al. Melatonin pretreatment alleviates the long-term synaptic toxicity and dysmyelination induced by neonatal sevoflurane exposure via MT1 receptor-mediated Wnt signaling modulation[J]. J Pineal Res, 2021, 71(4): e12771. |
| 9 | RAPER J, DE BIASIO J C, MURPHY K L, et al. Persistent alteration in behavioural reactivity to a mild social stressor in rhesus monkeys repeatedly exposed to sevoflurane in infancy[J]. Br J Anaesth, 2018, 120(4): 761-767. |
| 10 | YIN J, ZHAO X, WANG L J, et al. Sevoflurane-induced inflammation development: involvement of cholinergic anti-inflammatory pathway[J]. Behav Pharmacol, 2019, 30(8): 730-737. |
| 11 | SUN M Y, XIE Z C, ZHANG J Q, et al. Mechanistic insight into sevoflurane-associated developmental neurotoxicity[J]. Cell Biol Toxicol, 2022, 38(6): 927-943. |
| 12 | XIE L H, LIU Y, HU Y H, et al. Neonatal sevoflurane exposure induces impulsive behavioral deficit through disrupting excitatory neurons in the medial prefrontal cortex in mice[J]. Transl Psychiatry, 2020, 10(1): 202. |
| 13 | XU X Y, TIAN X, WANG G L. Sevoflurane reduced functional connectivity of excitatory neurons in prefrontal cortex during working memory performance of aged rats[J]. Biomed Pharmacother, 2018, 106: 1258-1266. |
| 14 | ZHAO T Y, CHEN Y X, SUN Z X, et al. Prenatal sevoflurane exposure causes neuronal excitatory/inhibitory imbalance in the prefrontal cortex and neurofunctional abnormality in rats[J]. Neurobiol Dis, 2020, 146: 105121. |
| 15 | ZHANG L, CHENG Y Y, XUE Z Y, et al. Sevoflurane impairs m6A-mediated mRNA translation and leads to fine motor and cognitive deficits[J]. Cell Biol Toxicol, 2022, 38(2): 347-369. |
| 16 | JIANG J L, LI S S, WANG Y Q, et al. Potential neurotoxicity of prenatal exposure to sevoflurane on offspring: metabolomics investigation on neurodevelopment and underlying mechanism[J]. Int J Dev Neurosci, 2017, 62: 46-53. |
| 17 | WANG C Y, LIU F, FRISCH-DAIELLO J L, et al. Lipidomics reveals a systemic energy deficient state that precedes neurotoxicity in neonatal monkeys after sevoflurane exposure[J]. Anal Chimica Acta, 2018, 1037: 87-96. |
| 18 | CHENG Y Y, LIU S Y, ZHANG L, et al. Identification of prefrontal cortex and amygdala expressed genes associated with sevoflurane anesthesia on non-human primate[J]. Front Integr Neurosci, 2022, 16: 857349. |
| 19 | XU G, LU H, DONG Y, et al. Coenzyme Q10 reduces sevoflurane-induced cognitive deficiency in young mice[J]. Br J Anaesth, 2017, 119(3): 481-491. |
| 20 | ZHANG J, DONG Y L, ZHOU C, et al. Anesthetic sevoflurane reduces levels of hippocalcin and postsynaptic density protein 95[J]. Mol Neurobiol, 2015, 51(3): 853-863. |
| 21 | ZHANG L, XUE Z Y, LIU Q D, et al. Disrupted folate metabolism with anesthesia leads to myelination deficits mediated by epigenetic regulation of ERMN[J]. EBioMedicine, 2019, 43: 473-486. |
| 22 | CHEN S F, ZHOU Y Q, CHEN Y R, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-i890. |
| 23 | LIM L, MI D, LLORCA A, et al. Development and functional diversification of cortical interneurons[J]. Neuron, 2018, 100(2): 294-313. |
| 24 | HAMMOND T R, DUFORT C, DISSING-OLESEN L, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes[J]. Immunity, 2019, 50(1): 253-271.e6. |
| 25 | JOGLEKAR A, PRJIBELSKI A, MAHFOUZ A, et al. A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain[J]. Nat Commun, 2021, 12(1): 463. |
| 26 | LI X S, LIU G P, YANG L, et al. Decoding cortical glial cell development[J]. Neurosci Bull, 2021, 37(4): 440-460. |
| 27 | MARQUES S, ZEISEL A, CODELUPPI S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system[J]. Science, 2016, 352(6291): 1326-1329. |
| 28 | WOLF F A, ANGERER P, THEIS F J. SCANPY: large-scale single-cell gene expression data analysis[J]. Genome Biol, 2018, 19(1): 15. |
| 29 | AIBAR S, GONZÁLEZ-BLAS C B, MOERMAN T, et al. SCENIC: single-cell regulatory network inference and clustering[J]. Nat Methods, 2017, 14(11): 1083-1086. |
| 30 | ASHBURNER M, BALL C A, BLAKE J A, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium[J]. Nat Genet, 2000, 25(1): 25-29. |
| 31 | DRAGHICI S, KHATRI P, TARCA A L, et al. A systems biology approach for pathway level analysis[J]. Genome Res, 2007, 17(10): 1537-1545. |
| 32 | YAARI G, BOLEN C R, THAKAR J, et al. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations[J]. Nucleic Acids Res, 2013, 41(18): e170. |
| 33 | BUTT S J B, SOUSA V H, FUCCILLO M V, et al. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes[J]. Neuron, 2008, 59(5): 722-732. |
| 34 | GRATTON M O, TORBAN E, JASMIN S B, et al. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms[J]. Mol Cell Biol, 2003, 23(19): 6922-6935. |
| 35 | EUN B, CHO B, MOON Y, et al. Induction of neuronal apoptosis by expression of Hes6 via p53-dependent pathway[J]. Brain Res, 2010, 1313: 1-8. |
| 36 | KOLDAMOVA R, SCHUG J, LEFTEROVA M, et al. Genome-wide approaches reveal EGR1-controlled regulatory networks associated with neurodegeneration[J]. Neurobiol Dis, 2014, 63: 107-114. |
| 37 | WANG C, QIN L N, MIN Z Q, et al. SOX7 interferes with β-catenin activity to promote neuronal apoptosis[J]. Eur J Neurosci, 2015, 41(11): 1430-1437. |
| 38 | CHENG B H, CHEN J, BAI B, et al. Neuroprotection of apelin and its signaling pathway[J]. Peptides, 2012, 37(1): 171-173. |
| 39 | ZHANG N, SU Q P, ZHANG W X, et al. Neuroprotection of dexmedetomidine against propofol-induced neuroapoptosis partly mediated by PI3K/Akt pathway in hippocampal neurons of fetal rat[J]. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 2017, 18(9): 789-796. |
| 40 | XUE H, XU Y, WANG S, et al. Sevoflurane post-conditioning alleviates neonatal rat hypoxic-ischemic cerebral injury via Ezh2-regulated autophagy[J]. Drug Des Devel Ther, 2019, 13: 1691-1706. |
| 41 | YANG Q Z, YAN W J, LI X, et al. Activation of canonical Notch signaling pathway is involved in the ischemic tolerance induced by sevoflurane preconditioning in mice[J]. Anesthesiology, 2012, 117(5): 996-1005. |
| 42 | ZHANG Y H, GAO Q S, WU Z Y, et al. Sevoflurane postconditioning ameliorates neuronal migration disorder through Reelin/Dab1 and improves long-term cognition in neonatal rats after hypoxic-ischemic injury[J]. Neurotox Res, 2021, 39(5): 1524-1542. |
| 43 | CHAI D D, CHENG Y Y, SUN Y, et al. Multiple sevoflurane exposures during pregnancy inhibit neuronal migration by upregulating prostaglandin D2 synthase[J]. Int J Dev Neurosci, 2019, 78: 77-82. |
| 44 | WANG X, SHAN Y Y, TANG Z Y, et al. Neuroprotective effects of dexmedetomidine against isoflurane-induced neuronal injury via glutamate regulation in neonatal rats[J]. Drug Des Devel Ther, 2018, 13: 153-160. |
| 45 | CHLEILAT E, SKATULLA L, RAHHAL B, et al. TGF-β signaling regulates development of midbrain dopaminergic and hindbrain serotonergic neuron subgroups[J]. Neuroscience, 2018, 381: 124-137. |
| 46 | JIANG M, TANG T X, LIANG X Y, et al. Maternal sevoflurane exposure induces temporary defects in interkinetic nuclear migration of radial glial progenitors in the fetal cerebral cortex through the Notch signalling pathway[J]. Cell Prolif, 2021, 54(6): e13042. |
| 47 | POON C L, MITCHELL K A, KONDO S, et al. The Hippo pathway regulates neuroblasts and brain size in Drosophila melanogaster[J]. Curr Biol, 2016, 26(8): 1034-1042. |
| 48 | GALDERISI U, JORI F P, GIORDANO A. Cell cycle regulation and neural differentiation[J]. Oncogene, 2003, 22(33): 5208-5219. |
| 49 | LIU S W, FANG F, SONG R X, et al. Sevoflurane affects neurogenesis through cell cycle arrest via inhibiting Wnt/β-catenin signaling pathway in mouse neural stem cells[J]. Life Sci, 2018, 209: 34-42. |
| 50 | SONG S Y, PENG K, MENG X W, et al. Single-nucleus atlas of sevoflurane-induced hippocampal cell type-and sex-specific effects during development in mice[J]. Anesthesiology, 2023, 138(5): 477-495. |
| [1] | 梁乐斌, 陈慧芳, 赖淑静, 顾靓, 苏冰. 基于空间ATAC-seq技术的Apcmin/+小鼠结肠肿瘤表观特征分析[J]. 上海交通大学学报(医学版), 2025, 45(10): 1261-1270. |
| [2] | 施灵玲, 程燕咏, 张磊. 七氟烷对原代少突胶质细胞增殖和分化的影响[J]. 上海交通大学学报(医学版), 2024, 44(9): 1115-1123. |
| [3] | 倪红伟1,贺广宝1,高红梅1,祝义军1,史东平1,杭燕南2. 困难气道的风险因素分析及预测模型研究[J]. 上海交通大学学报(医学版), 2020, 40(3): 358-. |
| [4] | 蒋云凤,程燕咏,孙宇. 长链非编码 RNA Peg13在七氟烷所致的发育期原代神经元凋亡中的作用[J]. 上海交通大学学报(医学版), 2019, 39(5): 469-. |
| [5] | 李璞玉,王 振. 重复经颅磁刺激在强迫症治疗中的应用[J]. 上海交通大学学报(医学版), 2019, 39(12): 1477-. |
| [6] | 刘配配,郑吉建,白洁,张马忠,孙瑛. 采用灌注指数监测小儿全身麻醉期间伤害性刺激反应的临床观察[J]. 上海交通大学学报(医学版), 2018, 38(1): 67-. |
| [7] | 仲 勇,周 嘉,袁 岚. 两种不同麻醉方式对患者围手术期应激的影响[J]. 上海交通大学学报(医学版), 2013, 33(12): 1681-. |
| [8] | 沈耀峰, 吴镜湘, 徐美英. 丙泊酚和七氟烷麻醉对老年患者普通胸外科手术后认知功能的影响[J]. , 2011, 31(3): 322-. |
| [9] | 张箐箐. 全身麻醉药物对小儿及幼年动物中枢神经系统影响的研究进展[J]. , 2010, 30(2): 233-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
