
Journal of Shanghai Jiao Tong University (Medical Science) ›› 2023, Vol. 43 ›› Issue (10): 1227-1235.doi: 10.3969/j.issn.1674-8115.2023.10.002
• Basic research • Previous Articles Next Articles
LIU Junjun1,2( ), LU Sumei2, ZHANG Bingyang2, LI Yongqing2, MA Wanshan1,2(
), LU Sumei2, ZHANG Bingyang2, LI Yongqing2, MA Wanshan1,2( )
)
Received:2023-04-14
Accepted:2023-09-12
Online:2023-10-28
Published:2023-10-28
Contact:
MA Wanshan
E-mail:liujing920927@163.com;mwsqianyi@163.com
Supported by:CLC Number:
LIU Junjun, LU Sumei, ZHANG Bingyang, LI Yongqing, MA Wanshan. Analysis of m6A methylation expression profiles in liver tissue of high-fat diet-induced mouse models of NAFLD[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(10): 1227-1235.
Add to citation manager EndNote|Ris|BibTeX
URL: https://xuebao.shsmu.edu.cn/EN/10.3969/j.issn.1674-8115.2023.10.002
| Gene | mRNA_ID | Locus | RNA length/nt | Fold change | P value |
|---|---|---|---|---|---|
| Up-regulated gene | |||||
| Micalc1 | ENSMUST00000033033 | chr7: 112374345-112395355: + | 2 259 | 2.840 | 0.004 |
| Cyp2b10 | ENSMUST00000005477 | chr7: 25897676-25926559: + | 1 822 | 2.721 | 0.011 |
| Ksr2 | ENSMUST00000180430 | chr5: 117414000-117775003: + | 13 125 | 2.484 | 0.001 |
| Ybx1 | ENSMUST00000079644 | chr4: 119277981-119294604: - | 1 628 | 2.359 | 0.023 |
| Paqr7 | ENSMUST00000095074 | chr4: 134497004-134510235: + | 3 550 | 2.274 | 0.002 |
| Casc5 | ENSMUST00000099542 | chr2: 119047119-119104121: + | 6 525 | 2.270 | 0.000 |
| Ubr3 | ENSMUST00000131553 | chr2: 69897303-69938659: + | 1 801 | 2.113 | 0.005 |
| Dpm3 | ENSMUST00000107462 | chr3: 89259358-89267077: + | 827 | 1.978 | 0.025 |
| Cd36 | ENSMUST00000082367 | chr5: 17782016-17835696: - | 3 016 | 1.965 | 0.021 |
| Tsc22d1 | ENSMUST00000142683 | chr14: 76487759-76506998: + | 797 | 1.928 | 0.028 |
| Down-regulated gene | |||||
| Esd | ENSMUST00000176957 | chr14: 74732384-74749848: + | 1 117 | 0.356 | 0.010 |
| Slc22a26 | ENSMUST00000120522 | chr19: 7781041-7802578: - | 2 854 | 0.421 | 0.010 |
| Folr1 | NM_001252553 | chr7: 101858331-101870788: - | 1 337 | 0.437 | 0.000 |
| Slc46a3 | ENSMUST00000138244 | chr5: 147893881-147894815: - | 388 | 0.437 | 0.001 |
| Inpp5d | ENSMUST00000167032 | chr1: 87676239-87720502: + | 4 125 | 0.462 | 0.010 |
| Hoxd9 | ENSMUST00000059272 | chr2: 74697727-74700208: + | 2 136 | 0.471 | 0.012 |
| Romo1 | ENSMUST00000088610 | chr2: 156144039-156145797: + | 569 | 0.484 | 0.005 |
| Litaf | ENSMUST00000117360 | chr16: 10960824-10975579: - | 763 | 0.492 | 0.014 |
| Mdh2 | ENSMUST00000019323 | chr5: 135778480-135790391: + | 1 456 | 0.509 | 0.032 |
| Sycp3 | ENSMUST00000125612 | chr10: 88459664-88473236: + | 1 169 | 0.513 | 0.044 |
Tab 1 Top 10 differentially m6A-methylated mRNAs
| Gene | mRNA_ID | Locus | RNA length/nt | Fold change | P value |
|---|---|---|---|---|---|
| Up-regulated gene | |||||
| Micalc1 | ENSMUST00000033033 | chr7: 112374345-112395355: + | 2 259 | 2.840 | 0.004 |
| Cyp2b10 | ENSMUST00000005477 | chr7: 25897676-25926559: + | 1 822 | 2.721 | 0.011 |
| Ksr2 | ENSMUST00000180430 | chr5: 117414000-117775003: + | 13 125 | 2.484 | 0.001 |
| Ybx1 | ENSMUST00000079644 | chr4: 119277981-119294604: - | 1 628 | 2.359 | 0.023 |
| Paqr7 | ENSMUST00000095074 | chr4: 134497004-134510235: + | 3 550 | 2.274 | 0.002 |
| Casc5 | ENSMUST00000099542 | chr2: 119047119-119104121: + | 6 525 | 2.270 | 0.000 |
| Ubr3 | ENSMUST00000131553 | chr2: 69897303-69938659: + | 1 801 | 2.113 | 0.005 |
| Dpm3 | ENSMUST00000107462 | chr3: 89259358-89267077: + | 827 | 1.978 | 0.025 |
| Cd36 | ENSMUST00000082367 | chr5: 17782016-17835696: - | 3 016 | 1.965 | 0.021 |
| Tsc22d1 | ENSMUST00000142683 | chr14: 76487759-76506998: + | 797 | 1.928 | 0.028 |
| Down-regulated gene | |||||
| Esd | ENSMUST00000176957 | chr14: 74732384-74749848: + | 1 117 | 0.356 | 0.010 |
| Slc22a26 | ENSMUST00000120522 | chr19: 7781041-7802578: - | 2 854 | 0.421 | 0.010 |
| Folr1 | NM_001252553 | chr7: 101858331-101870788: - | 1 337 | 0.437 | 0.000 |
| Slc46a3 | ENSMUST00000138244 | chr5: 147893881-147894815: - | 388 | 0.437 | 0.001 |
| Inpp5d | ENSMUST00000167032 | chr1: 87676239-87720502: + | 4 125 | 0.462 | 0.010 |
| Hoxd9 | ENSMUST00000059272 | chr2: 74697727-74700208: + | 2 136 | 0.471 | 0.012 |
| Romo1 | ENSMUST00000088610 | chr2: 156144039-156145797: + | 569 | 0.484 | 0.005 |
| Litaf | ENSMUST00000117360 | chr16: 10960824-10975579: - | 763 | 0.492 | 0.014 |
| Mdh2 | ENSMUST00000019323 | chr5: 135778480-135790391: + | 1 456 | 0.509 | 0.032 |
| Sycp3 | ENSMUST00000125612 | chr10: 88459664-88473236: + | 1 169 | 0.513 | 0.044 |
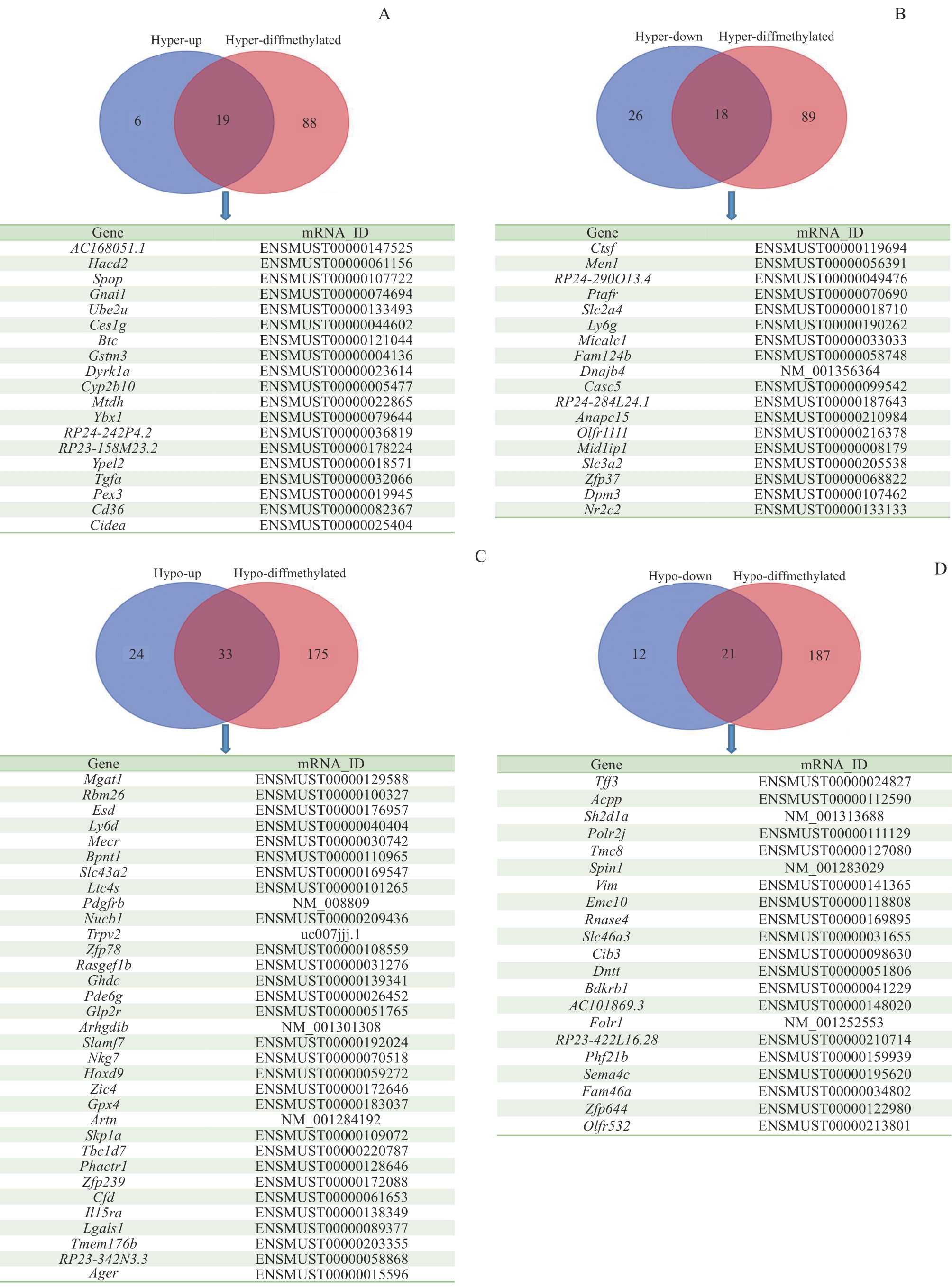
Fig 4 Differential m6A modification and differentially expressed genes in mouse liver tissueNote: A to D—Venn diagrams for high/low m6A methylation and high/low expression genes.
| 1 | 祥蔚. IL-17A基因敲除减轻LPS诱导的脂肪肝小鼠炎症性肝损伤及其机制研究[D].重庆: 第三军医大学, 2017. |
| XIANG W. IL-17A deficiency alleviates LPS-induced steatotic liver injury in mice and the underlying mechanism[D]. Chongqing: Third Military Medical University of Chinese P.L.A., 2017. | |
| 2 | ESTES C, ANSTEE Q M, ARIAS-LOSTE M T, et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016‒2030[J]. J Hepatol, 2018, 69(4): 896-904. |
| 3 | CHENG Y, HOU T, PING J, et al. Quantitative succinylome analysis in the liver of non-alcoholic fatty liver disease rat model[J]. Proteome Sci, 2016, 14: 3. |
| 4 | XU Z J, SHI J P, YU D R, et al. Evaluating the relationship between metabolic syndrome and liver biopsy-proven non-alcoholic steatohepatitis in China: a multicenter cross-sectional study design[J]. Adv Ther, 2016, 33(11): 2069-2081. |
| 5 | YOUNOSSI Z M, KOENIG A B, ABDELATIF D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes[J]. Hepatology, 2016, 64(1): 73-84. |
| 6 | YOUNOSSI Z M, GOLABI P, DE AVILA L, et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis[J]. J Hepatol, 2019, 71(4): 793-801. |
| 7 | PASCHOS P, PALETAS K. Non alcoholic fatty liver disease and metabolic syndrome[J]. Hippokratia, 2009, 13(1): 9-19. |
| 8 | STEFAN N, HÄRING H U, CUSI K. Non-alcoholic fatty liver disease: causes, diagnosis, cardiometabolic consequences, and treatment strategies[J]. Lancet Diabetes Endocrinol, 2019, 7(4): 313-324. |
| 9 | ALEXANDER M, LOOMIS A K, VAN DER LEI J, et al. Risks and clinical predictors of cirrhosis and hepatocellular carcinoma diagnoses in adults with diagnosed NAFLD: real-world study of 18 million patients in four European cohorts[J]. BMC Med, 2019, 17(1): 95. |
| 10 | FRIEDMAN S L, NEUSCHWANDER-TETRI B A, RINELLA M, et al. Mechanisms of NAFLD development and therapeutic strategies[J]. Nat Med, 2018, 24(7): 908-922. |
| 11 | DIEHL A M, DAY C. Cause, pathogenesis, and treatment of nonalcoholic steatohepatitis[J]. N Engl J Med, 2017, 377(21): 2063-2072. |
| 12 | 王园园, 蒋建萍. 非酒精性脂肪肝与代谢综合征各组分关系的研究进展[J]. 中国当代医药, 2022, 29(15): 36-39, 43. |
| WANG Y Y, JIANG J P. Research progress of the relationship between nonalcoholic fatty liver disease and components of metabolic syndrome[J]. China Modern Medicine, 2022, 29(15): 36-39, 43. | |
| 13 | ZHOU J, ZHOU F, WANG W, et al. Epidemiological features of NAFLD from 1999 to 2018 in China[J]. Hepatology, 2020, 71(5): 1851-1864. |
| 14 | 李永清. METTL3介导的CYP2B6 m6A甲基化修饰在非酒精性脂肪性肝病肝细胞胰岛素敏感性中的作用和机制研究[D]. 济南: 山东大学, 2022. |
| LI Y Q. Role and mechanism of METTL3-mediated CYP2B6 m6A methylation modification in insulin sensitivity in hepatocytes with non-alcoholic fatty liver disease[D]. Jinan: Shandong University, 2022. | |
| 15 | 叶棣文, 马万山, 逯素梅. RNAm6A甲基化修饰在代谢综合征中的研究进展[J]. 临床检验杂志, 2022, 40(9): 691-694. |
| YE D W, MA W S, LU S M. Advances in the study of RNA m6A methylation modification in the metabolic syndrome[J].Chinese Journal of Clinical Laboratory Science, 2022,40(9): 691-694. | |
| 16 | SUMIDA Y, YONEDA M. Current and future pharmacological therapies for NAFLD/NASH[J]. J Gastroenterol, 2018, 53(3): 362-376. |
| 17 | 陈子扬, 蒲锐, 邓爽, 等. m6A甲基化与代谢性疾病调控[J]. 中国组织工程研究, 2022, 26(20): 3250-3255. |
| CHEN Z Y, PU R, DENG S, et al. N6-methyladenosine methylation and its regulation in metabolic diseases[J]. Chinese Journal of Tissue Engineering Research, 2022, 26(20):3250-3255. | |
| 18 | PENG Z, GONG Y, WANG X, et al. METTL3-m6A-Rubicon axis inhibits autophagy in nonalcoholic fatty liver disease[J]. Mol Ther, 2022, 30(2): 932-946. |
| 19 | 韦冷云. Hacd2在脂质代谢中的功能及其作用机制[D]. 无锡: 江南大学, 2022. |
| WEI L Y. Function and mechanism of Hacd2 in lipid metabolism[D]. Wuxi: Jiangnan University, 2022. | |
| 20 | 曾晗. CD36在肝细胞脂质从头合成中的作用及机制研究[D]. 重庆: 重庆医科大学, 2022. |
| ZENG H. The role of CD36 in regulation of hepatocyte de novo lipogenesis[D]. Chongqing: ChongQing Medical University, 2022. | |
| 21 | 仝雪薇, 冶学燕, 刘春燕, 等. 2型糖尿病合并肥胖患者外周血PPARG、CIDEA、ECHDC3、CGN mRNA表达与血脂水平的关系研究[J]. 深圳中西医结合杂志, 2021, 31(6): 4-8, 199. |
| TONG X W, YE X Y, LIU C Y, et al. Relationship between mRNA expression of PPARG, CIDEA, ECHDC3, CGN and lipid levels in peripheral blood of patients with type 2 diabetes mellitus complicated with obesity[J].Shenzhen Journal of Integrated Traditional Chinese and Western Medicine, 2021, 31(6): 4-8, 199. | |
| 22 | SANS A, BONNAFOUS S, ROUSSEAU D, et al. The differential expression of CIDE family members is associated with NAFLD progression from steatosis to steatohepatitis[J]. Sci Rep, 2019, 9(1): 7501. |
| 23 | TANG J, ZHAO X, WEI W, et al. METTL16-mediated translation of CIDEA promotes non-alcoholic fatty liver disease progression via m6A-dependent manner[J]. PeerJ, 2022, 10: e14379. |
| 24 | QUIROGA A D, LI L, TRÖTZMÜLLER M, et al. Deficiency of carboxylesterase 1/esterase-x results in obesity, hepatic steatosis, and hyperlipidemia[J]. Hepatology, 2012, 56(6): 2188-2198. |
| 25 | 钱胜南. ATP在胰岛素抵抗机制中对线粒体MECR蛋白的影响[D]. 上海: 上海海洋大学, 2020. |
| QIAN S N. Effect of ATP on mitochondrial MECR protein in the mechanism of insulin resistance[D].Shanghai: Shanghai Ocean University, 2020. | |
| 26 | 潘晓燕. FOXO转录因子在饮食诱导的脂肪性肝病中的作用机制研究[D]. 苏州: 苏州大学, 2018. |
| PAN X Y. Study on the mechanisms of action of FOXO transcriptional factors in diet-induced fatty liver disease[D].Suzhou: Soochow University, 2018. |
| [1] | SONG Jing, JIANG Shuo, WAN Fangyu, LI Juan, MUHETA Adina, MIN Xinying, ZHOU Jingqi. Research progress in effects and mechanisms of dietary pattern interventions in metabolic associated fatty liver disease [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(7): 926-933. |
| [2] | BIAN Shu, YU Qian, LIU Liangming. Research progress in the role of endo cannabinoid system in liver diseases [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(10): 1299-1306. |
| [3] | BENEDICK Jun Er Chin, SON Peng, ZHANG Yifan, WANG Junqing, GUO Simin. Research progress of the impact of nonalcoholic fatty liver disease on chronic hepatitis B infection [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(12): 1585-1590. |
| [4] | SHEN Haiqian, YU Kangying, CHEN Yingying, JI Ping, WANG Ying. Construction of an mRNA vaccine encoding hemagglutinin of influenza A H1N1 virus and investigation on booster immunization strategy [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(11): 1374-1383. |
| [5] | ZONG Chunyan, HE Jie, ZHANG Zhe, JIA Renbing, SHEN Jianfeng. Role of APOBEC3B in regulating replication stress of uveal melanoma [J]. Journal of Shanghai Jiao Tong University (Medical Science), 2022, 42(8): 1034-1044. |
| [6] | Xiaowen ZHANG, Yi WANG, Chan ZHANG, Di ZHANG, Hang YUN, Di HUANG. Effects of Pcsk9 gene interference on high fat-induced nonalcoholic fatty liver disease with atherosclerosis in rats [J]. JOURNAL OF SHANGHAI JIAOTONG UNIVERSITY (MEDICAL SCIENCE), 2022, 42(2): 150-157. |
| [7] | Hai-hong WANG, Shuo GAO, Jun ZHU, Jun ZHOU. Expression patterns of interferon regulatory factor 2 binding protein 2 during the early embryonic development of zebrafish [J]. JOURNAL OF SHANGHAI JIAOTONG UNIVERSITY (MEDICAL SCIENCE), 2021, 41(3): 314-319. |
| [8] | Chao SANG, Dan-dan LIANG, Guo-xiang XIE, Wei JIA, Tian-lu CHEN. Progress in serological noninvasive diagnostic methods for non-alcoholic fatty liver disease [J]. JOURNAL OF SHANGHAI JIAOTONG UNIVERSITY (MEDICAL SCIENCE), 2021, 41(1): 112-117. |
| [9] | LIU Yong-mei, TAN Ming, LEI Ming. Electron microscopic study of the human THOC1-THOC2-THOC3 subcomplex [J]. , 2020, 40(1): 1-. |
| [10] | YING Chen1, LIU Cai-hong2, HU Jia-an2, JIANG Shi-hu3, XU Zhi-hong2, SUN Jing2. Research on level of serum lipid metabolism related hormones in patients of obstructive sleep apnoea hypoxia syndrome combine with non-alcoholic fatty liver disease [J]. , 2018, 38(10): 1203-. |
| [11] | FAN Meng-xia, QIAO Jie. Research progresses of luteinizing hormone receptor mRNA binding protein [J]. , 2015, 35(6): 906-. |
| [12] | ZHANG Sheng-zhi, SHAO Hua-jiang, MA Jian-ting. Advances of relationship between E6/E7 mRNA of human papillomavirus and cervical lesions [J]. , 2014, 34(5): 754-. |
| [13] | XU Ming, XU Jie-li, ZHOU Ying-zi, et al. Value of differential diagnosis of hTERT mRNA, UbcH10 and Ki-67 in breast apocrine lesions [J]. , 2013, 33(9): 1239-. |
| [14] | LU Qin-chi, CAI Guo-en, CHEN Sheng-di. Expression of metabotropic glutamate receptor 1, 4, 7 in CA1 region of hippocampus in pilocarpine-induced temporal lobe epilepsy [J]. , 2010, 30(4): 394-. |
| [15] | ZHANG Peng, WU Ju-gang, WU Hong-biao, et al. Expression of CD133 mRNA in tissues of gastric cancer and its relationship with clinicopathological features [J]. , 2010, 30(2): 213-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||