
上海交通大学学报(医学版) ›› 2023, Vol. 43 ›› Issue (11): 1396-1407.doi: 10.3969/j.issn.1674-8115.2023.11.007
• 论著 · 基础研究 • 上一篇
赵富茂1( ), 彭玫1, 彭晓露1, 舒韦韦2, 彭丽1(
), 彭玫1, 彭晓露1, 舒韦韦2, 彭丽1( )
)
收稿日期:2023-02-28
接受日期:2023-09-19
出版日期:2023-11-28
发布日期:2023-11-28
通讯作者:
彭丽
E-mail:506884746@qq.com;pli1228@163.com
作者简介:赵富茂(1995—),女,硕士生;电子信箱:506884746@qq.com。
基金资助:
ZHAO Fumao1( ), PENG Mei1, PENG Xiaolu1, SHU Weiwei2, PENG Li1(
), PENG Mei1, PENG Xiaolu1, SHU Weiwei2, PENG Li1( )
)
Received:2023-02-28
Accepted:2023-09-19
Online:2023-11-28
Published:2023-11-28
Contact:
PENG Li
E-mail:506884746@qq.com;pli1228@163.com
Supported by:摘要:
目的·探寻鲍曼不动杆菌(Acinetobacter baumannii)在环境中碳青霉烯类药物美罗培南浓度改变时耐药性变化的机制。方法·通过改变鲍曼不动杆菌标准敏感株ATCC19606和临床耐药株AB.2014培养环境中的美罗培南浓度等条件,诱导对美罗培南不同耐药程度的衍生株。测量所得菌株的生长曲线,并提取各菌株的DNA和RNA,采用PCR分析菌株耐药性改变后的碳青霉烯酶基因IMI、KPC、GES-1、IMP、VIM、NDM-1、OXA23、OXA24、OXA51、OXA58的表达情况;通过实时荧光定量PCR(real-time fluorescent quantitative PCR,RT-qPCR)分析不同耐药程度的鲍曼不动杆菌耐药基因,包括OXA51,外排泵基因adeB、adeG、adeJ,孔蛋白基因carO、omp33-36、oprC,青霉素结合蛋白基因ponA的表达水平变化;通过全基因组测序及生物信息学工具分析耐药性改变后菌株的差异基因富集情况的变化。结果·获得了鲍曼不动杆菌ATCC19606与AB.2014对美罗培南不同耐药程度的11个衍生株,最低抑菌浓度(minimum inhibitory concentration,MIC)为1~128 μg/mL。ATCC19606及其衍生株的生长速度和峰值随着耐药性的增加而降低,但AB.2014及其衍生株并没有表现出这种趋势。ATCC19606及其衍生株表达3个碳青霉烯酶基因OXA51、VIM和IMP,AB.2014及其多数衍生株表达4个碳青霉烯酶基因OXA23、OXA51、VIM和IMP,仅AB.2014的一个复敏衍生株出现了OXA23丢失。RT-qPCR结果显示,仅在ATCC19606及其耐药衍生株中oprC基因的表达量随着耐药性的升高而降低,多数耐药基因的表达水平与菌株的耐药水平变化一致。生物信息学分析提示ATCC19606不同衍生株之间的差异基因主要富集于铁载体摄取跨膜转运体活性、细胞外膜、细菌分泌系统和群体感应等,而AB.2014不同衍生株之间的差异基因主要富集于细胞外膜、细胞对化学刺激的反应、阿特拉津降解和RNA聚合酶等。结论·碳青霉烯类药物环境压力会引起鲍曼不动杆菌耐药性发生变化,碳青霉烯酶、外排泵、孔蛋白、青霉素结合蛋白多种基因可能同时参与了菌株耐药性的变化;碳青霉烯酶OXA23丢失可能导致耐药鲍曼不动杆菌对碳青霉烯类药物复敏。
| Ambler class | Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) | Fragment size/bp | Reference |
|---|---|---|---|---|---|
| A | IMI | ATAGCCATCCTTGTTTAGCTC | TCTGCGATTACTTTATCCTC | 818 | [ |
| KPC | GCTTCCCACTGTGCAGCTCATTCA | CGGTCGTGTTTCCCTTTAGCCAATC | 456 | - | |
| GES-1 | ATGCGCTTCATTCACGCAC | CTATTTGTCCGTGCTAAGG | 814 | [ | |
| B | IMP | AGGCCAAGATCGTTTAGTCACTGT | GTGTTGTACAGATAGAACCAGGACCAA | 461 | - |
| VIM | TGGTGAGTATCCGACAGTCAACGA | CGAATGCGCAGCACCAG | 450 | - | |
| NDM-1 | AGGACAAGATGGGCGGTATGGACG | CCATGCGGGCCGTATGAGTGATT | 429 | - | |
| D | OXA51 | ATGAACATTAAAGCACTC | CTATAAAATACCTAATTGTTC | 825 | [ |
| OXA23 | ACTTGCTATGTGGTTGCTTC | TGGAAGCTGTGTATGTGCTA | 555 | [ | |
| OXA24 | TTTGCCGATGACCTTGCACATAAC | TCATGTTGAGCGAAAAGGGGATTTTT | 208 | - | |
| OXA58 | CGATCAGAATGTTCAAGCGC | TCCCCTCTGCGCTCTACATACA | 666 | - |
表1 碳青霉烯酶基因PCR鉴定引物
Tab 1 Primers used in the identification of carbapenemase genes by PCR
| Ambler class | Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) | Fragment size/bp | Reference |
|---|---|---|---|---|---|
| A | IMI | ATAGCCATCCTTGTTTAGCTC | TCTGCGATTACTTTATCCTC | 818 | [ |
| KPC | GCTTCCCACTGTGCAGCTCATTCA | CGGTCGTGTTTCCCTTTAGCCAATC | 456 | - | |
| GES-1 | ATGCGCTTCATTCACGCAC | CTATTTGTCCGTGCTAAGG | 814 | [ | |
| B | IMP | AGGCCAAGATCGTTTAGTCACTGT | GTGTTGTACAGATAGAACCAGGACCAA | 461 | - |
| VIM | TGGTGAGTATCCGACAGTCAACGA | CGAATGCGCAGCACCAG | 450 | - | |
| NDM-1 | AGGACAAGATGGGCGGTATGGACG | CCATGCGGGCCGTATGAGTGATT | 429 | - | |
| D | OXA51 | ATGAACATTAAAGCACTC | CTATAAAATACCTAATTGTTC | 825 | [ |
| OXA23 | ACTTGCTATGTGGTTGCTTC | TGGAAGCTGTGTATGTGCTA | 555 | [ | |
| OXA24 | TTTGCCGATGACCTTGCACATAAC | TCATGTTGAGCGAAAAGGGGATTTTT | 208 | - | |
| OXA58 | CGATCAGAATGTTCAAGCGC | TCCCCTCTGCGCTCTACATACA | 666 | - |
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) | Reference |
|---|---|---|---|
| 16S rRNA | GTAGCTTGCTACTGGACCTAG | CATACTCTAGCTCACCAGTATCG | [ |
| OXA51 | GATTTAGCTCGTCGTATTGGA | AAGCGTTTTATTAGCTAGCTTG | [ |
| adeB | GGAATAAGGCACCACAACAAT | CGAAGTTAGGAATACCAGCAATAC | - |
| adeG | TCACCAGATAATCGCTATG | GACTTCACCTACACCTTG | - |
| adeJ | CCTATTGCACAATATCCAACGA | AGGATAAGTCGCAGCAATCG | [ |
| oprC | ACTCGATACAAAGCGGTGGA | TTTAATACGTGAACCAAACATACCTC | [ |
| carO | TGTTCATGACAGCTATGCATTCGATA | CCCAATGCTAAACCTACATATGGGT | [ |
| omp33-36 | GCAACTTACAACCACACTGA | TAACAACATAGCACCAACTTCTAA | - |
| ponA | GTCAGCCAGGTTCTACCATCAA | CCATCAGAGTTCTTCGGTGTCC | - |
表2 RT-qPCR引物
Tab 2 Primers used in RT-qPCR analysis
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) | Reference |
|---|---|---|---|
| 16S rRNA | GTAGCTTGCTACTGGACCTAG | CATACTCTAGCTCACCAGTATCG | [ |
| OXA51 | GATTTAGCTCGTCGTATTGGA | AAGCGTTTTATTAGCTAGCTTG | [ |
| adeB | GGAATAAGGCACCACAACAAT | CGAAGTTAGGAATACCAGCAATAC | - |
| adeG | TCACCAGATAATCGCTATG | GACTTCACCTACACCTTG | - |
| adeJ | CCTATTGCACAATATCCAACGA | AGGATAAGTCGCAGCAATCG | [ |
| oprC | ACTCGATACAAAGCGGTGGA | TTTAATACGTGAACCAAACATACCTC | [ |
| carO | TGTTCATGACAGCTATGCATTCGATA | CCCAATGCTAAACCTACATATGGGT | [ |
| omp33-36 | GCAACTTACAACCACACTGA | TAACAACATAGCACCAACTTCTAA | - |
| ponA | GTCAGCCAGGTTCTACCATCAA | CCATCAGAGTTCTTCGGTGTCC | - |
| Strain | MIC/(μg·mL-1) |
|---|---|
| ATCC19606 | 1 |
| ATCC19606-R1 | 8 |
| ATCC19606-R2 | 16 |
| ATCC19606-R3 | 128 |
| ATCC19606-R3-S1 | 64 |
| ATCC19606-R3-S2 | 1 |
| AB.2014 | 16 |
| AB.2014-R1 | 64 |
| AB.2014-R2 | 128 |
| AB.2014-S1 | 8 |
| AB.2014-S2 | 2 |
| AB.2014-R1-S1 | 16 |
| AB.2014-R1-S2 | 8 |
表3 ATCC19606、AB.2014及它们衍生株对MEM的MIC值
Tab 3 MIC values of ATCC19606, AB.2014 and their derivative strains to MEM
| Strain | MIC/(μg·mL-1) |
|---|---|
| ATCC19606 | 1 |
| ATCC19606-R1 | 8 |
| ATCC19606-R2 | 16 |
| ATCC19606-R3 | 128 |
| ATCC19606-R3-S1 | 64 |
| ATCC19606-R3-S2 | 1 |
| AB.2014 | 16 |
| AB.2014-R1 | 64 |
| AB.2014-R2 | 128 |
| AB.2014-S1 | 8 |
| AB.2014-S2 | 2 |
| AB.2014-R1-S1 | 16 |
| AB.2014-R1-S2 | 8 |
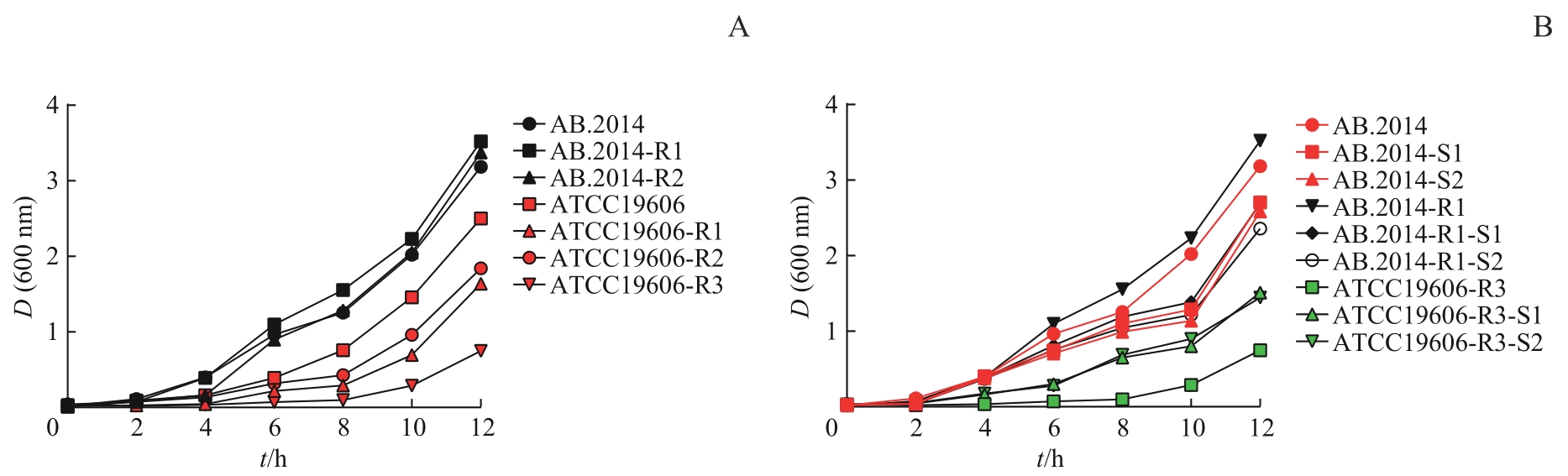
图1 不同耐药性鲍曼不动杆菌菌株的生长曲线Note: A. Growth curves of the strains with increased MEM-resistance. B. Growth curves of the strains with decreased MEM-resistance.
Fig 1 Growth curves of A. baumannii strains with different drug resistance
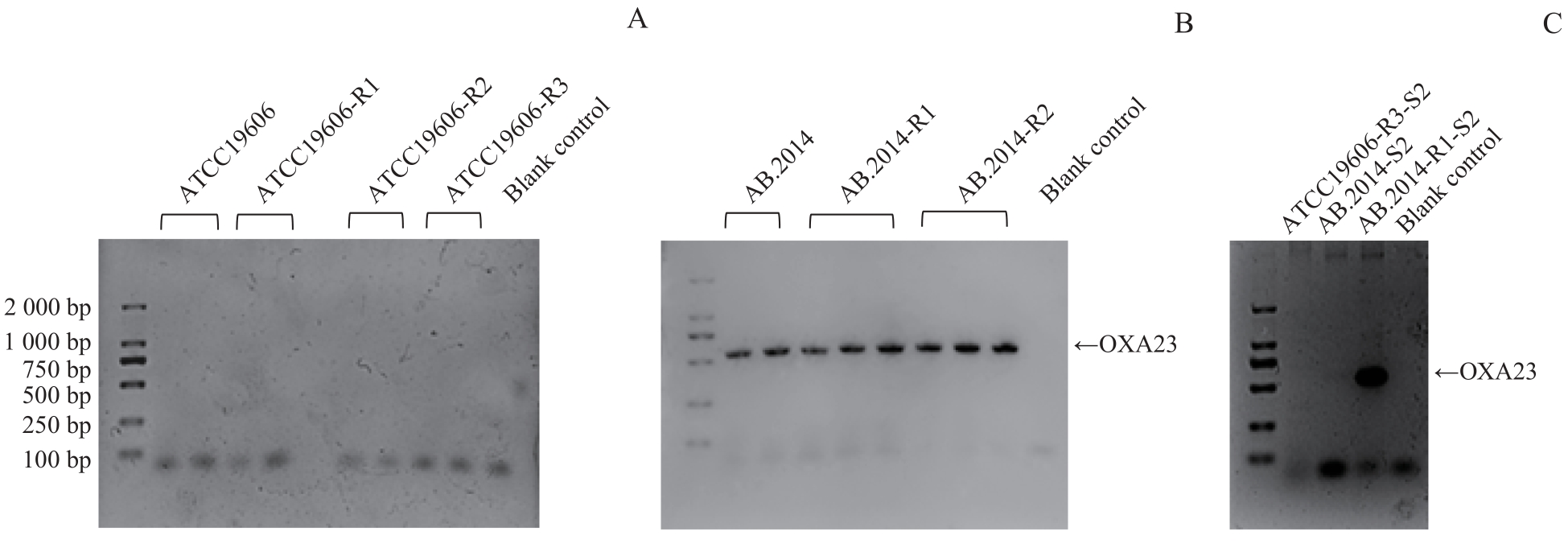
图2 不同耐药性鲍曼不动杆菌菌株中 OXA23 基因的PCR鉴定Note: A. Detection of OXA23 gene expression in ATCC19606 and its MEM-resistant variants. B.Detection of OXA23 gene expression in AB.2014 and its MEM-resistant variants. C.Detection of OXA23 gene expression in MEM-sensitive variants.
Fig 2 Analysis of OXA23 gene expression in A. baumannii strains with different drug resistance by PCR
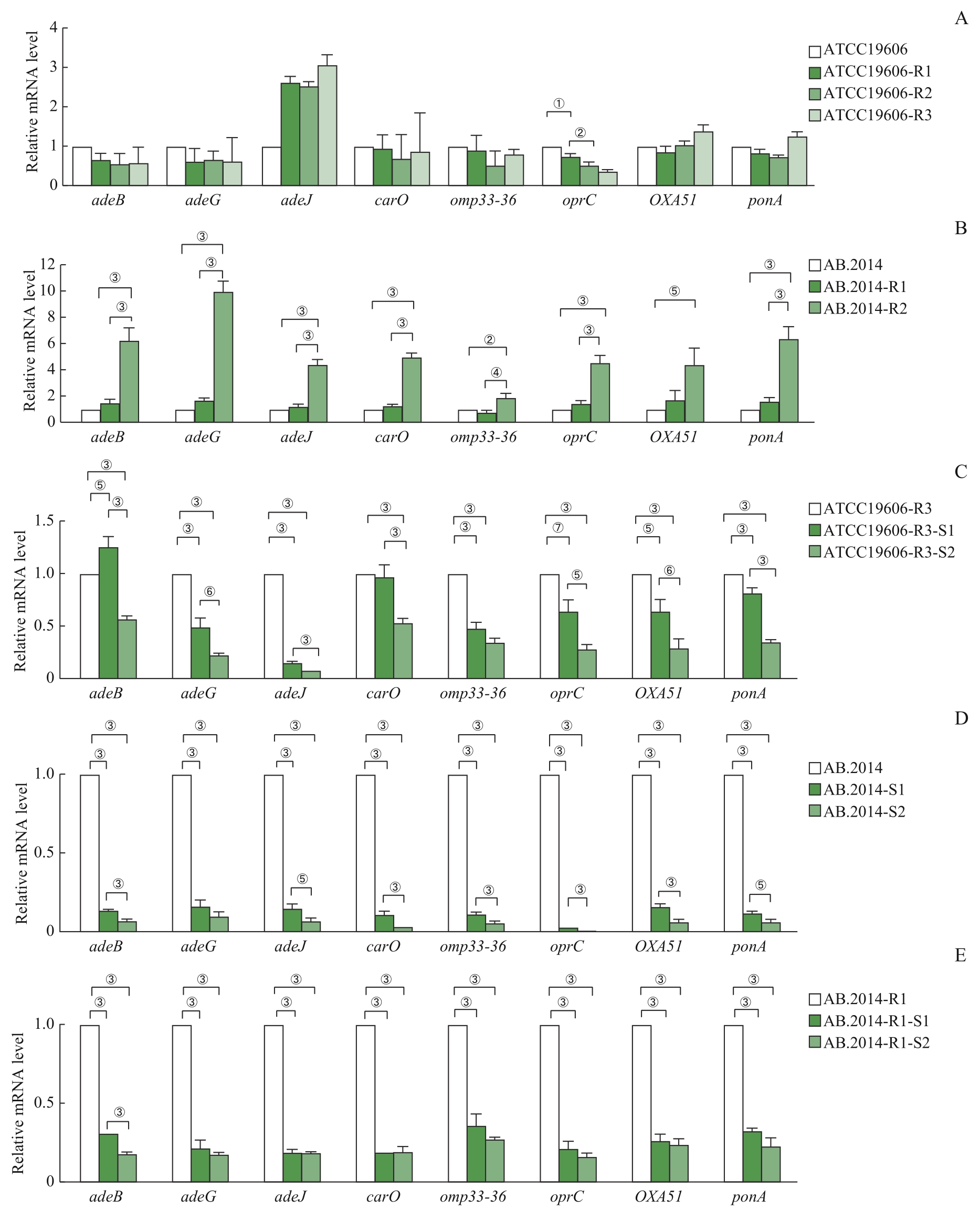
图3 RT-qPCR检测不同耐药性鲍曼不动杆菌菌株中抗MEM相关基因的表达Note: A. Expression profiles of the genes in ATCC19606 and its MEM-resistant variants. B. Expression profiles of the genes in AB.2014 and its MEM-resistant variants. C. Expression profiles of the genes in ATCC19606-R3 and its MEM-sensitive variants. D. Expression profiles of the genes in AB.2014 and its MEM-sensitive variants. E. Expression profiles of the genes of AB.2014-R1 and its MEM-sensitive variants. ①P=0.005, ②P= 0.003, ③P=0.000, ④P=0.007, ⑤P=0.001, ⑥P=0.002, ⑦P=0.006.
Fig 3 RT-qPCR analysis of MEM-resistance-related genes in A. baumannii strains with different drug resistance
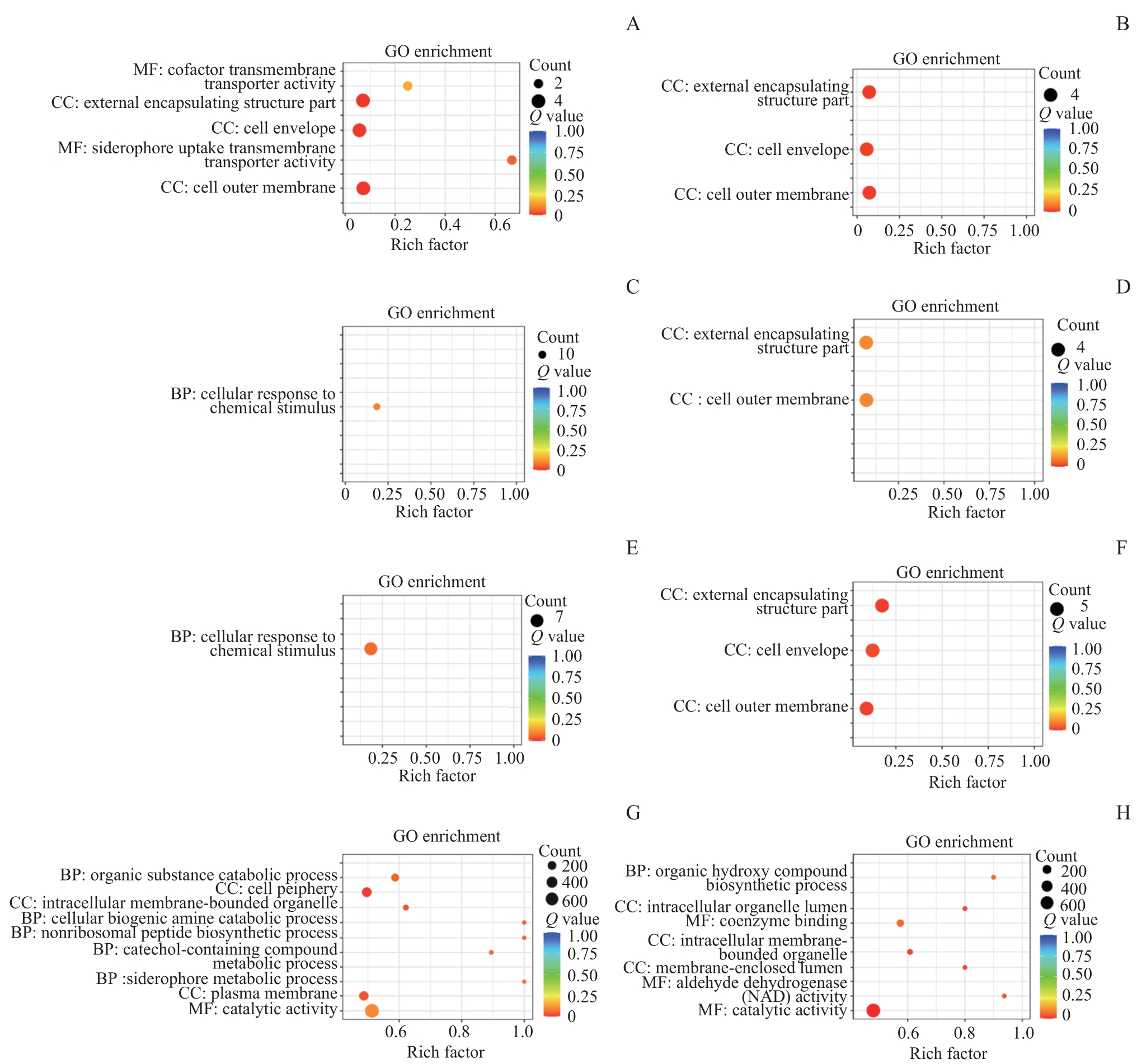
图4 ATCC19606、AB.2014及它们衍生株之间差异基因的GO分析Note: A. Enrichment of differential genes in ATCC19606 compared with ATCC19606-R3. B. Enrichment of differential genes in ATCC19606-R3-S2 compared with ATCC19606-R3. C. Enrichment of differential genes in AB.2014 compared with AB.2014-R3. D. Enrichment of differential genes in AB.2014-R3 compared with AB.2014. E. Enrichment of differential genes in AB.2014 compared with AB.2014-S2. F. Enrichment of differential genes in AB.2014-S2 compared with AB.2014. G. Enrichment of differential genes in ATCC19606 compared with AB.2014. H. Enrichment of differential genes in AB.2014 compared with ATCC19606.
Fig 4 GO analysis of differential genes among ATCC19606, AB.2014 and their derivatives
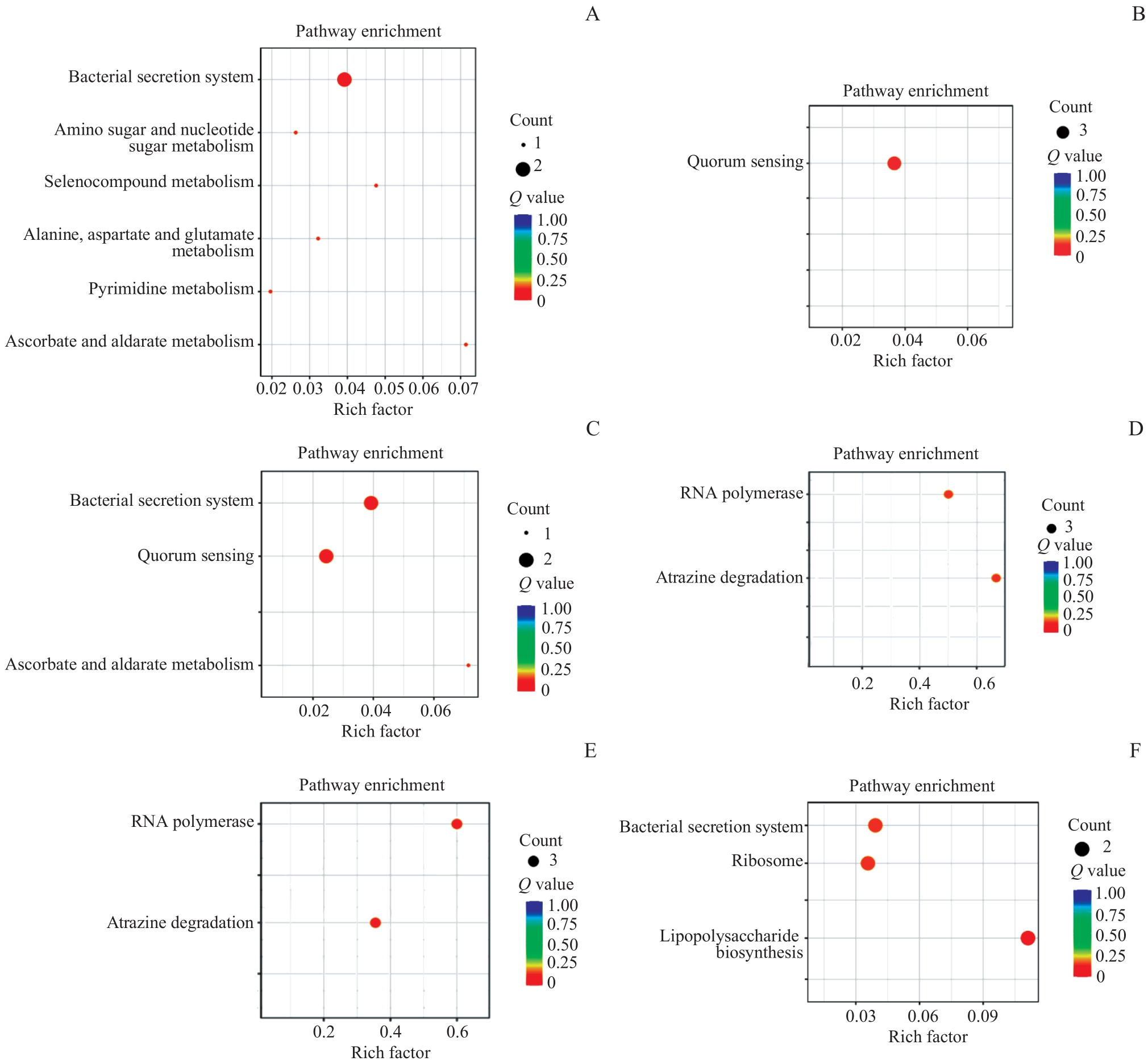
图5 ATCC19606、AB.2014及它们衍生株之间差异基因的KEGG分析Note: A. Enrichment of differential genes in ATCC19606 compared with ATCC19606-R3. B. Enrichment of differential genes in ATCC19606-R3 compared with ATCC19606. C. Enrichment of differential genes in ATCC19606-R3 compared with ATCC19606-R3-S2. D. Enrichment of differential genes in AB.2014 compared with AB.2014-R3. E. Enrichment of differential genes in AB.2014 compared with AB.2014-S2. F. Enrichment of differential genes in AB.2014-R2-S2 compared with AB.2014-R2.
Fig 5 KEGG analysis of differential genes among ATCC19606, AB.2014 and their derivatives
| 1 | DOURAGHI M, KENYON J J, ARIS P, et al. Accumulation of antibiotic resistance genes in carbapenem-resistant Acinetobacter baumannii isolates belonging to lineage 2, global clone 1, from outbreaks in 2012-2013 at a Tehran burns hospital[J]. mSphere, 2020, 5(2): e00164-e00120. |
| 2 | RAMIREZ M S, BONOMO R A, TOLMASKY M E. Carbapenemases: transforming Acinetobacter baumannii into a yet more dangerous menace[J]. Biomolecules, 2020, 10(5): 720. |
| 3 | MENG X, FU J T, ZHENG Y, et al. Ten-year changes in bloodstream infection with Acinetobacter baumannii complex in intensive care units in Eastern China: a retrospective cohort study[J]. Front Med (Lausanne), 2021, 8: 715213. |
| 4 | HAMIDIAN M, NIGRO S J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii[J]. Microb Genom, 2019, 5(10): e000306. |
| 5 | VÁZQUEZ-UCHA J C, ARCA-SUÁREZ J, BOU G, et al. New carbapenemase inhibitors: clearing the way for the β-lactams[J]. Int J Mol Sci, 2020, 21(23): 9308. |
| 6 | CHOQUET M, LOHOU E, PAIR E, et al. Efflux pump overexpression profiling in Acinetobacter baumannii and study of new 1-(1-naphthylmethyl)-piperazine analogs as potential efflux inhibitors[J]. Antimicrob Agents Chemother, 2021, 65(9): e0071021. |
| 7 | UPPALAPATI S R, SETT A, PATHANIA R. The outer membrane proteins OmpA, CarO, and OprD of Acinetobacter baumannii confer a two-pronged defense in facilitating its success as a potent human pathogen[J]. Front Microbiol, 2020, 11: 589234. |
| 8 | HERNÁNDEZ-ROCAMORA V M, BARANOVA N, PETERS K, et al. Real-time monitoring of peptidoglycan synthesis by membrane-reconstituted penicillin-binding proteins[J]. eLife, 2021, 10: e61525. |
| 9 | BOLL J M, CROFTS A A, PETERS K, et al. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii[J]. Proc Natl Acad Sci USA, 2016, 113(41): E6228-E6237. |
| 10 | MOSTAFAVI S N, KHEDMATI M, KELISHADI R. Microbiology and antimicrobial sensitivity of ventriculo-peritoneal shunt infections in a referral paediatric neurosurgery ward during a period of 7 years[J]. J Glob Antimicrob Resist, 2022, 29: 63-67. |
| 11 | CHEN X, MENG X B, GAO Q Q, et al. Meropenem selection induced overproduction of the intrinsic carbapenemase as well as phenotype divergence in Acinetobacter baumannii[J]. Int J Antimicrob Agents, 2017, 50(3): 419-426. |
| 12 | AUBRON C, POIREL L, ASH R J, et al. Carbapenemase-producing Enterobacteriaceae, U.S. rivers[J]. Emerg Infect Dis, 2005, 11(2): 260-264. |
| 13 | SHAHCHERAGHI F, NIKBIN V S, FEIZABADI M M. Prevalence of ESBLs genes among multidrug-resistant isolates of Pseudomonas aeruginosa isolated from patients in Tehran[J]. Microb Drug Resist, 2009, 15(1): 37-39. |
| 14 | ZHOU S R, CHEN X, MENG X B, et al. “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii[J]. Sci Rep, 2015, 5: 8976. |
| 15 | KUO H Y, CHANG K C, KUO J W, et al. Imipenem: a potent inducer of multidrug resistance in Acinetobacter baumannii[J]. Int J Antimicrob Agents, 2012, 39(1): 33-38. |
| 16 | HU W S, YAO S M, FUNG C P, et al. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii[J]. Antimicrob Agents Chemother, 2007, 51(11): 3844-3852. |
| 17 | RUMBO C, GATO E, LÓPEZ M, et al. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii[J]. Antimicrob Agents Chemother, 2013, 57(11): 5247-5257. |
| 18 | SEWE S O, SILVA G, SICAT P, et al. Trimming and validation of illumina short reads using trimmomatic, trinity assembly, and assessment of RNA-seq data[J]. Methods Mol Biol, 2022, 2443: 211-232. |
| 19 | ULINTZ P J, WU W S, GATES C M. Bioinformatics analysis of whole exome sequencing data[J]. Methods Mol Biol, 2019, 1881: 277-318. |
| 20 | WU T Z, HU E Q, XU S B, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data[J]. Innovation (Camb), 2021, 2(3): 100141. |
| 21 | KNOPP M, ANDERSSON D I. Amelioration of the fitness costs of antibiotic resistance due to reduced outer membrane permeability by upregulation of alternative porins[J]. Mol Biol Evol, 2015, 32(12): 3252-3263. |
| 22 | DA SILVA K E, MACIEL W G, CRODA J, et al. A high mortality rate associated with multidrug-resistant Acinetobacter baumannii ST79 and ST25 carrying OXA-23 in a Brazilian intensive care unit[J]. PLoS One, 2018, 13(12): e0209367. |
| 23 | BLACKWELL G A, HALL R M. Mobilisation of a small Acinetobacter plasmid carrying an oriT transfer origin by conjugative RepAci6 plasmids[J]. Plasmid, 2019, 103: 36-44. |
| 24 | BARTSCH A, IVES C M, KATTNER C, et al. An antibiotic-resistance conferring mutation in a neisserial porin: structure, ion flux, and ampicillin binding[J]. Biochim Biophys Acta Biomembr, 2021, 1863(6): 183601. |
| 25 | BHAMIDIMARRI S P, YOUNG T R, SHANMUGAM M, et al. Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site[J]. PLoS Biol, 2021, 19(11): e3001446. |
| 26 | ZHANG M L, CHEN L H, YE C S, et al. Co-selection of antibiotic resistance via copper shock loading on bacteria from a drinking water bio-filter[J]. Environ Pollut, 2018, 233: 132-141. |
| 27 | SCOFFONE V C, TRESPIDI G, BARBIERI G, et al. Role of RND efflux pumps in drug resistance of cystic fibrosis pathogens[J]. Antibiotics (Basel), 2021, 10(7): 863. |
| 28 | TOTH M, LEE M, STEWART N K, et al. Effects of inactivation of D,D-transpeptidases of Acinetobacter baumannii on bacterial growth and susceptibility to β-lactam antibiotics[J]. Antimicrob Agents Chemother, 2022, 66(1): e0172921. |
| 29 | YI L, DONG X, GRENIER D, et al. Research progress of bacterial quorum sensing receptors: classification, structure, function and characteristics[J]. Sci Total Environ, 2021, 763: 143031. |
| 30 | VALASTYAN J S, KRAML C M, PELCZER I, et al. Saccharomyces cerevisiae requires CFF1 to produce 4-hydroxy-5-methylfuran-3(2H)- one, a mimic of the bacterial quorum-sensing autoinducer AI-2[J]. mBio, 2021, 12(2): e03303-e03320. |
| [1] | 陆文清, 孟周文理, 虞永峰, 陆舜. 非小细胞肺癌第三代表皮生长因子受体-酪氨酸激酶抑制剂的耐药机制及治疗策略[J]. 上海交通大学学报(医学版), 2022, 42(4): 535-544. |
| [2] | 徐建华, 江萍, 邓炯. ATP结合盒蛋白G超家族成员2在肺癌中的表达及意义[J]. 上海交通大学学报(医学版), 2021, 41(6): 830-833. |
| [3] | 郝艳云, 俞思慧, 陆静, 顾湘, 张帆, 程金科, 王田实. SIRT3去SUMO化修饰调节乳腺癌细胞MCF7增殖及化疗药物敏感性的研究[J]. 上海交通大学学报(医学版), 2021, 41(12): 1557-1563. |
| [4] | 王钰婷1, 2,刘锦燕2,史 册1,赵珺涛1, 2,项明洁1, 2. 白念珠菌ERG3基因敲除及其对耐药性的影响[J]. 上海交通大学学报(医学版), 2020, 40(2): 163-. |
| [5] | 余春波 1,卢明 2,邵雷 3,蒲甜 4,陈代杰 4,周薇 1, 5. relA基因敲除对多黏菌素抗鲍曼不动杆菌异质性耐药的影响[J]. 上海交通大学学报(医学版), 2019, 39(9): 1004-. |
| [6] | 吕 霖,石 鑫,郭晓奎,秦金红. 耐药肺炎克雷伯菌噬菌体JD902的分离及生物学特性和安全性研究[J]. 上海交通大学学报(医学版), 2019, 39(12): 1389-. |
| [7] | 季佳炜 1, 2,王睿 1,罗婷婷 4,徐梦莎 1,郭晓奎 1,胡付品 3,李敏 2,何平 1. 耐碳青霉烯类鲍曼不动杆菌噬菌体SH-Ab15599的生物学特性及基因组学分析[J]. 上海交通大学学报(医学版), 2018, 38(11): 1283-. |
| [8] | 刘洋,郑丹丹,韩逸超,史玮炀,戴尔宽,李敏,郑冰 . 多重耐药肺炎克雷伯菌感染的危险因素及治疗方案比较[J]. 上海交通大学学报(医学版), 2017, 37(7): 973-. |
| [9] | 肖淑珍,韩立中,王文奎. 279株泌尿生殖道分离解脲脲原体的药敏分析[J]. 上海交通大学学报(医学版), 2016, 36(4): 546-. |
| [10] | 李文静,刘锦燕,史册,等. 融合PCR结合同源重组技术敲除白色假丝酵母菌FLO8基因[J]. 上海交通大学学报(医学版), 2016, 36(3): 334-. |
| [11] | 刘锦燕,史册,王影,等. 白假丝酵母菌唑类耐药相关的转录调控研究进展[J]. 上海交通大学学报(医学版), 2016, 36(2): 291-. |
| [12] | 刘婧娴,俞静,李媛睿,等. 肺炎克雷伯菌对碳青霉烯类抗生素的耐药机制研究[J]. 上海交通大学学报(医学版), 2016, 36(1): 93-. |
| [13] | 黄洁,顾秋莹,李梅玲,等. 外科重症监护病房多重耐药鲍曼不动杆菌交叉感染的防控效果和临床特征分析[J]. 上海交通大学学报(医学版), 2015, 35(9): 1315-. |
| [14] | 向军,郭海娜,郇京宁. 烧伤临床鲍曼不动杆菌生物膜形成对外排泵调节基因adeS和adeR及其功能基因簇adeAB表达的影响[J]. 上海交通大学学报(医学版), 2015, 35(9): 1364-. |
| [15] | 赵静,王凤平,孙清清,等. DNA微阵列法检测结核分枝杆菌对利福平和异烟肼的耐药性[J]. 上海交通大学学报(医学版), 2015, 35(11): 1651-. |
| 阅读次数 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
全文 2376
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
摘要 486
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||