
上海交通大学学报(医学版) ›› 2025, Vol. 45 ›› Issue (2): 138-149.doi: 10.3969/j.issn.1674-8115.2025.02.002
邹沛辰1( ), 刘鸿宇2, 阿衣娜扎尔·艾合买提1, 朱亮1, 唐亚斌1(
), 刘鸿宇2, 阿衣娜扎尔·艾合买提1, 朱亮1, 唐亚斌1( ), 雷绘敏1(
), 雷绘敏1( )
)
收稿日期:2024-07-09
接受日期:2024-10-23
出版日期:2025-02-28
发布日期:2025-02-28
通讯作者:
唐亚斌,高级实验师,博士;电子信箱:leonyabin2018@shsmu.edu.cn作者简介:邹沛辰(1997—),男,硕士生;电子信箱:peichenzou1997@163.com。
基金资助:
ZOU Peichen1( ), LIU Hongyu2, AIHEMAITI· Ayinazhaer1, ZHU Liang1, TANG Yabin1(
), LIU Hongyu2, AIHEMAITI· Ayinazhaer1, ZHU Liang1, TANG Yabin1( ), LEI Huimin1(
), LEI Huimin1( )
)
Received:2024-07-09
Accepted:2024-10-23
Online:2025-02-28
Published:2025-02-28
Contact:
TANG Yabin, E-mail: leonyabin2018@shsmu.edu.cnSupported by:摘要:
目的·探究KRAS靶向抑制剂索托拉西布(sotorasib)获得性耐药肺癌细胞的代谢特征及代谢重编程规律。方法·构建肺癌细胞H2122和H358的索托拉西布获得性耐药细胞模型(H2122-SR和H358-SR细胞),并采用CCK-8法加以验证。应用超高效液相色谱串联四极杆飞行时间质谱(UPLC-QTOF/MS)分析获得性耐药肺癌细胞及其同源亲本细胞的代谢轮廓,利用主成分分析法和偏最小二乘法-判别分析法等统计学方法进行非靶向代谢组学分析及代谢表征,筛选并鉴定索托拉西布获得性耐药相关的差异代谢物,并对所筛选到的差异代谢物进行通路富集分析。通过绘制热图比较分析主要差异代谢通路上的代谢物在耐药细胞与亲本细胞之间的差异。结果·成功构建H2122和H358细胞的获得性耐药细胞模型,对索托拉西布的半数抑制浓度均较亲本细胞提升50倍以上。与亲本细胞相比,耐药细胞代谢轮廓存在显著差异。H2122-SR与H2122细胞之间发现48种差异代谢物,其中变量投影重要程度(variable importance in the projection,VIP)值排名前10位的差异代谢物为尿嘧啶、黄苷酸、吲哚-3-甲酸、烟酸、黄苷、黄嘌呤、N-甲基烟酰胺、次黄嘌呤、葫芦巴碱、半乳糖醛酸;H358-SR与H358细胞之间发现79种差异代谢物,其中VIP值排名前10位的差异代谢物为还原型谷胱甘肽、黄苷、α-酮戊二酸、羧甲基赖氨酸、胸苷、嘌呤、核黄素、3-吲哚丙烯酸、吲哚-3-丙酮酸、二氢尿嘧啶。2种肺癌细胞系的差异代谢通路主要涉及嘌呤代谢和糖酵解/糖异生,其中嘌呤代谢变化最为显著。热图分析显示,耐药细胞嘌呤代谢途径多数代谢物水平升高。结论·索托拉西布获得性耐药的肺癌细胞嘌呤代谢增强。
中图分类号:
邹沛辰, 刘鸿宇, 阿衣娜扎尔·艾合买提, 朱亮, 唐亚斌, 雷绘敏. 索托拉西布获得性耐药肺癌细胞的代谢轮廓分析[J]. 上海交通大学学报(医学版), 2025, 45(2): 138-149.
ZOU Peichen, LIU Hongyu, AIHEMAITI· Ayinazhaer, ZHU Liang, TANG Yabin, LEI Huimin. Metabolic profiling of lung cancer cells with acquired resistance to sotorasib[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(2): 138-149.
| Time/min | Flow rate/ (mL·min-1) | Mobile phase A/% | Mobile phase B/% |
|---|---|---|---|
| 0 | 0.25 | 10 | 90 |
| 1.5 | 0.25 | 10 | 90 |
| 20 | 0.25 | 60 | 40 |
| 25 | 0.25 | 60 | 40 |
| 25.01 | 0.25 | 10 | 90 |
| 33 | 0.25 | 10 | 90 |
表1 H2122-SR和H2122细胞样本的梯度洗脱程序
Tab 1 Gradient elution program for H2122-SR and H2122 cells
| Time/min | Flow rate/ (mL·min-1) | Mobile phase A/% | Mobile phase B/% |
|---|---|---|---|
| 0 | 0.25 | 10 | 90 |
| 1.5 | 0.25 | 10 | 90 |
| 20 | 0.25 | 60 | 40 |
| 25 | 0.25 | 60 | 40 |
| 25.01 | 0.25 | 10 | 90 |
| 33 | 0.25 | 10 | 90 |
| Time/min | Flow rate/ (mL·min-1) | Mobile phase A/% | Mobile phase B/% |
|---|---|---|---|
| 0 | 0.5 | 5 | 95 |
| 0.5 | 0.5 | 5 | 95 |
| 7 | 0.5 | 35 | 65 |
| 8 | 0.5 | 60 | 40 |
| 9 | 0.5 | 60 | 40 |
| 9.01 | 0.5 | 5 | 95 |
| 12 | 0.5 | 5 | 95 |
表2 H358-SR和H358细胞样本的梯度洗脱程序
Tab 2 Gradient elution program for H358-SR and H358 cells
| Time/min | Flow rate/ (mL·min-1) | Mobile phase A/% | Mobile phase B/% |
|---|---|---|---|
| 0 | 0.5 | 5 | 95 |
| 0.5 | 0.5 | 5 | 95 |
| 7 | 0.5 | 35 | 65 |
| 8 | 0.5 | 60 | 40 |
| 9 | 0.5 | 60 | 40 |
| 9.01 | 0.5 | 5 | 95 |
| 12 | 0.5 | 5 | 95 |
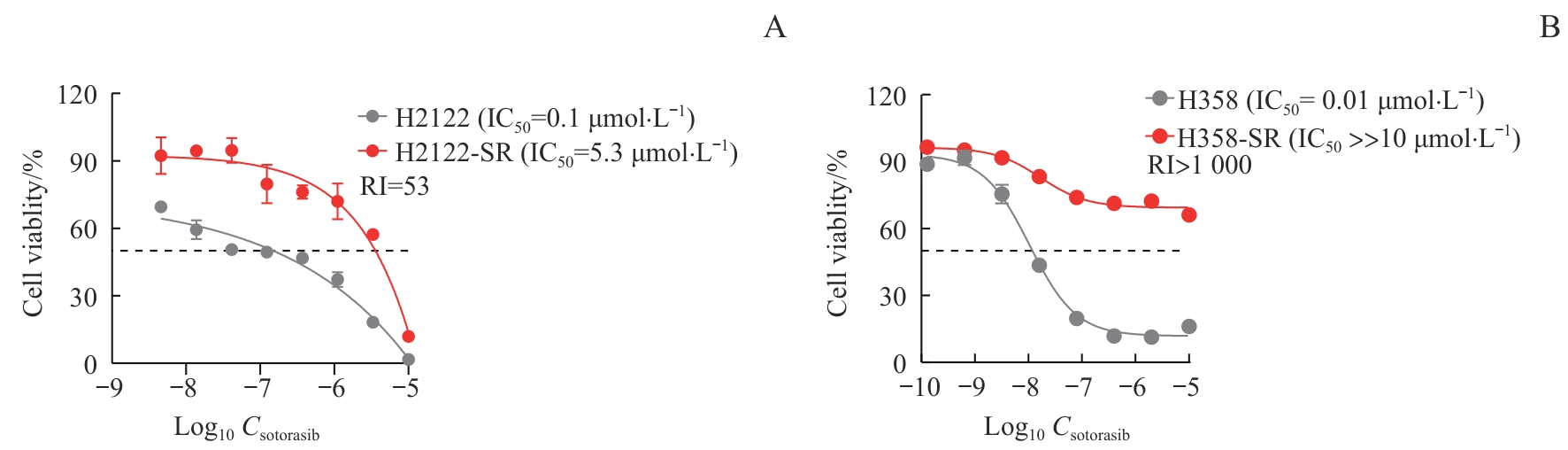
图1 索托拉西布耐药肺癌细胞模型的鉴定Note: The cells were treated with indicated concentrations of sotorasib for 72 h. The viability of sotorasib-resistant cells H2122-SR and H358-SR and their corresponding parental cells H2122 and H358 were analyzed by CCK-8 assay. A. H2122 and H2122-SR cells. B. H358 and H358-SR cells.
Fig 1 Identification of sotorasib-resistant cell models in lung cancer
图2 H2122和H2122-SR细胞代谢总离子流色谱图Note: A. Representative chromatogram of H2122 cells in ESI+. B. Representative chromatogram of H2122-SR cells in ESI+. C. Representative chromatogram of H2122 cells in ESI-. D. Representative chromatogram of H2122-SR cells in ESI-. cps—counts per second.
Fig 2 Representative chromatograms of H2122 and H2122-SR cells in ESI
图3 H358和H358-SR细胞代谢总离子流色谱图Note: A. Representative chromatogram of H358 cells in ESI+. B. Representative chromatogram of H358-SR cells in ESI+. C. Representative chromatogram of H358 cells in ESI-. D. Representative chromatogram of H358-SR cells in ESI-.
Fig 3 Representative chromatograms of H358 and H358-SR cells in ESI
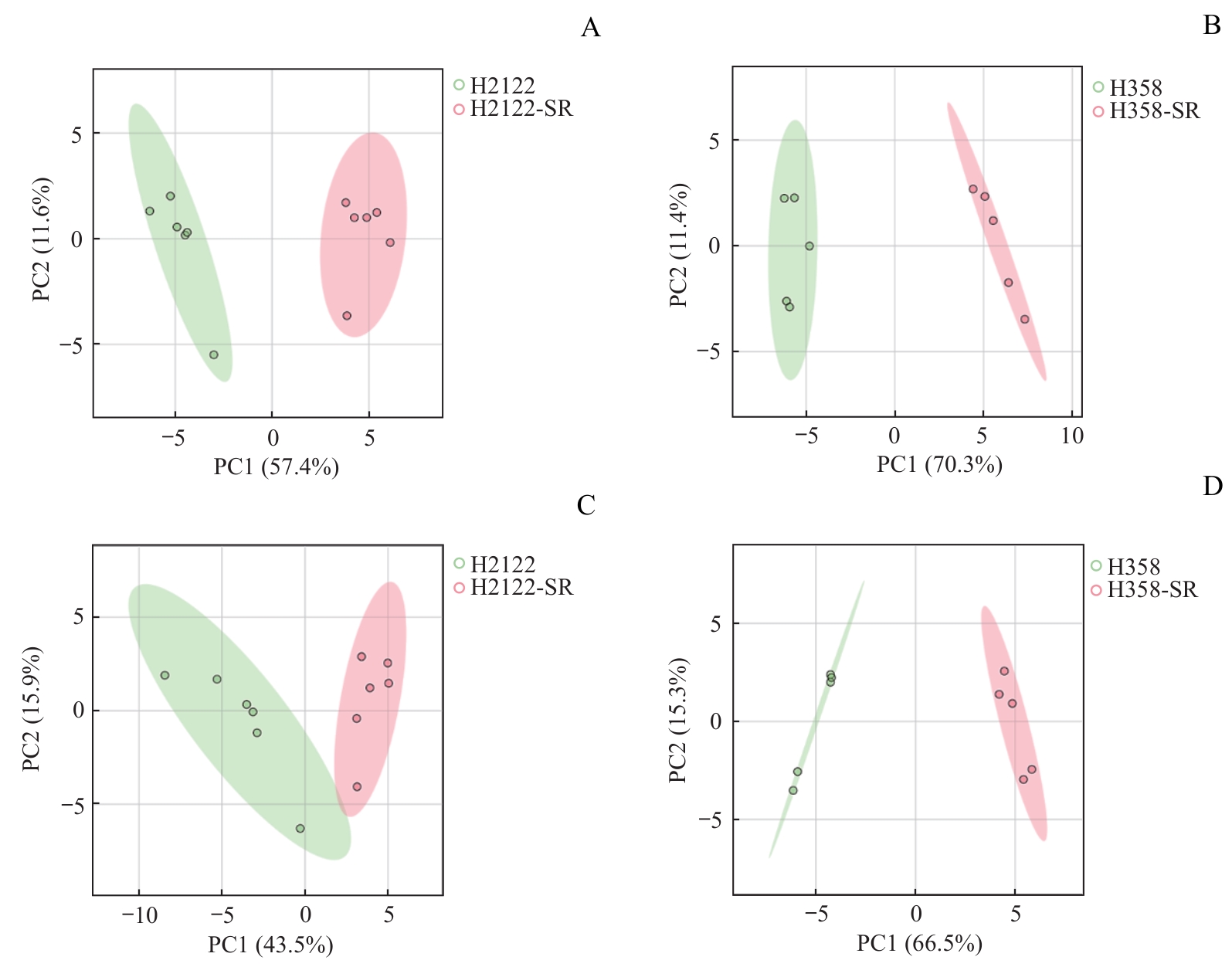
图4 H2122-SR和H358-SR细胞与其亲本细胞的PCA得分图Note: A/B. PCA score plots of H2122-SR (A, n=6) and H358-SR (B, n=5) with their corresponding parental cells in ESI+. C/D. PCA score plots of H2122-SR (C, n=6) and H358-SR (D, n=5) with their corresponding parental cells in ESI-. Green circles represent parental cells, and red circles represent SR cells.
Fig 4 PCA score plots of H2122-SR and H358-SR cells with their parental cells
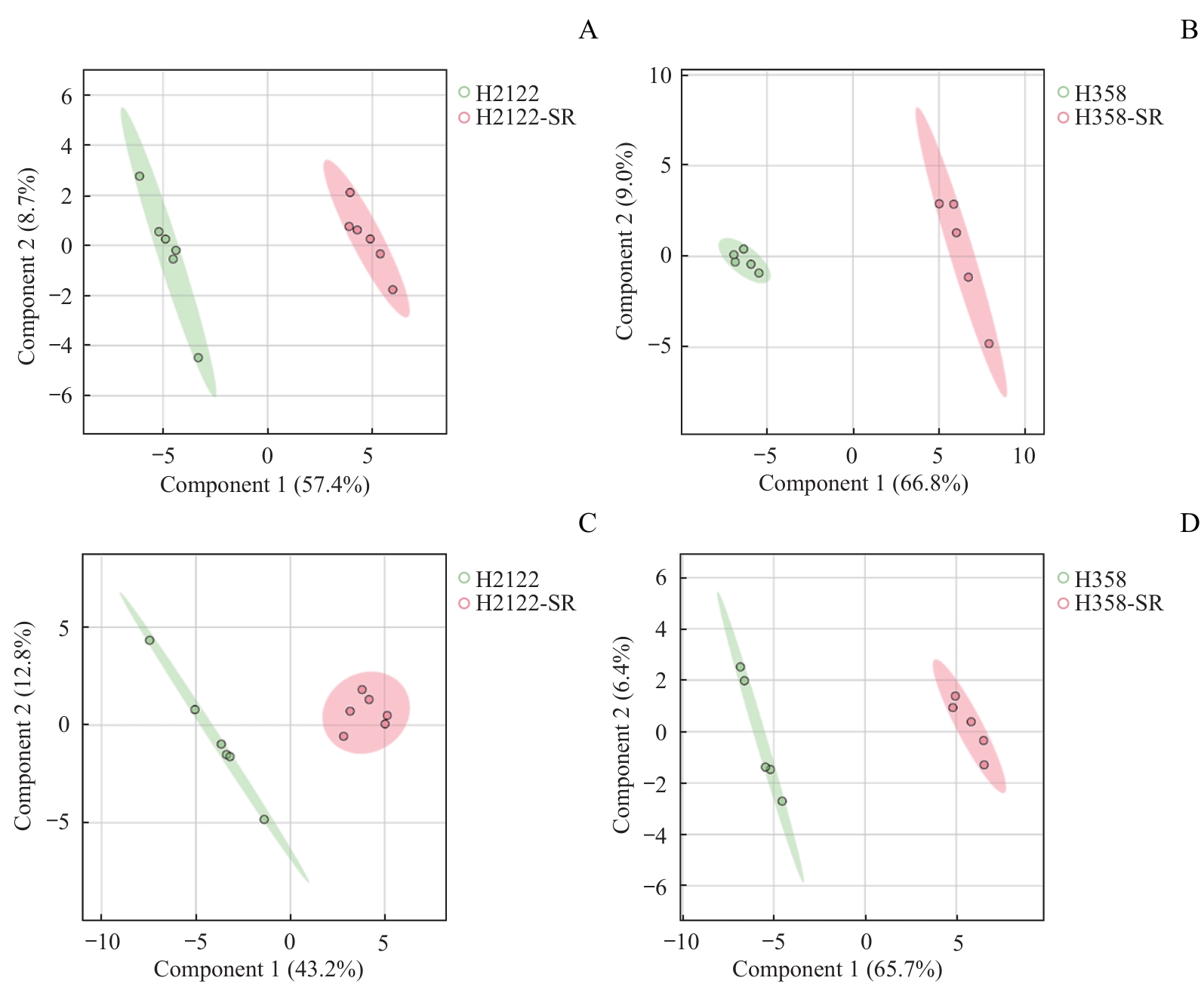
图5 H2122-SR和H358-SR细胞与其亲本细胞的PLS-DA得分图Note: A/B. PLS-DA score plots of H2122-SR (A, n=6) and H358-SR (B, n=5) with their corresponding parental cells in ESI+. C/D. PLS-DA score plots of H2122-SR (C, n=6) and H358-SR (D, n=5) with their corresponding parental cells in ESI-. Green circles represent parental cells, and red circles represent SR cells.
Fig 5 PLS-DA score plots of H2122-SR and H358-SR cells with their parental cells
| No. | Name | VIP | FC (SR/Par) | P value | Score |
|---|---|---|---|---|---|
| 1 | Uridine | 4.17 | 0.005 | 9.16×10-6 | 94 |
| 2 | Xanthylic acid | 3.02 | 25.748 | 1.19×10-5 | 96 |
| 3 | Indole-3-carboxylic acid | 2.80 | 18.718 | 7.79×10-8 | 100 |
| 4 | Nicotinic acid | 2.77 | 18.143 | 2.04×10-8 | 98 |
| 5 | Xanthosine | 2.42 | 8.841 | 2.17×10-6 | 93 |
| 6 | Xanthine | 2.29 | 7.136 | 2.48×10-6 | 99 |
| 7 | N-methylnicotinamide | 2.20 | 0.161 | 3.94×10-9 | 99 |
| 8 | Hypoxanthine | 2.14 | 5.898 | 3.33×10-6 | 99 |
| 9 | Trigonelline | 2.06 | 5.809 | 3.08×10-5 | 90 |
| 10 | Galactonic acid | 2.04 | 0.451 | 1.74×10-4 | 90 |
表3 H2122-SR与H2122细胞VIP排序前10位的差异代谢物
Tab 3 Top 10 differential metabolites in VIP ranking between H2122-SR and H2122 cells
| No. | Name | VIP | FC (SR/Par) | P value | Score |
|---|---|---|---|---|---|
| 1 | Uridine | 4.17 | 0.005 | 9.16×10-6 | 94 |
| 2 | Xanthylic acid | 3.02 | 25.748 | 1.19×10-5 | 96 |
| 3 | Indole-3-carboxylic acid | 2.80 | 18.718 | 7.79×10-8 | 100 |
| 4 | Nicotinic acid | 2.77 | 18.143 | 2.04×10-8 | 98 |
| 5 | Xanthosine | 2.42 | 8.841 | 2.17×10-6 | 93 |
| 6 | Xanthine | 2.29 | 7.136 | 2.48×10-6 | 99 |
| 7 | N-methylnicotinamide | 2.20 | 0.161 | 3.94×10-9 | 99 |
| 8 | Hypoxanthine | 2.14 | 5.898 | 3.33×10-6 | 99 |
| 9 | Trigonelline | 2.06 | 5.809 | 3.08×10-5 | 90 |
| 10 | Galactonic acid | 2.04 | 0.451 | 1.74×10-4 | 90 |
| No. | Name | VIP | FC (SR/Par) | P value | Score |
|---|---|---|---|---|---|
| 1 | Glutathione | 2.31 | 38.656 | 3.43×10-5 | 92 |
| 2 | Xanthosine | 2.25 | 31.638 | 4.47×10-7 | 85 |
| 3 | 2-Ketoglutaric acid | 2.21 | 25.561 | 1.64×10-4 | 99 |
| 4 | Carboxyethyl lysine | 2.11 | 38.608 | 3.73×10-4 | 90 |
| 5 | Thymidine | 2.10 | 0.036 | 1.05×10-4 | 85 |
| 6 | Purine | 2.05 | 18.272 | 3.43×10-5 | 90 |
| 7 | Riboflavin | 1.99 | 15.114 | 1.01×10-6 | 92 |
| 8 | 3-Indolylacrylic acid | 1.98 | 45.888 | 1.38×10-4 | 85 |
| 9 | Indole-3-pyruvic acid | 1.82 | 0.439 | 5.32×10-5 | 85 |
| 10 | Dihydrouracil | 1.79 | 15.319 | 6.22×10-7 | 100 |
表4 H358-SR与H358细胞VIP排序前10位的差异代谢物
Tab 4 Top 10 differential metabolites in VIP ranking between H358-SR and H358 cells
| No. | Name | VIP | FC (SR/Par) | P value | Score |
|---|---|---|---|---|---|
| 1 | Glutathione | 2.31 | 38.656 | 3.43×10-5 | 92 |
| 2 | Xanthosine | 2.25 | 31.638 | 4.47×10-7 | 85 |
| 3 | 2-Ketoglutaric acid | 2.21 | 25.561 | 1.64×10-4 | 99 |
| 4 | Carboxyethyl lysine | 2.11 | 38.608 | 3.73×10-4 | 90 |
| 5 | Thymidine | 2.10 | 0.036 | 1.05×10-4 | 85 |
| 6 | Purine | 2.05 | 18.272 | 3.43×10-5 | 90 |
| 7 | Riboflavin | 1.99 | 15.114 | 1.01×10-6 | 92 |
| 8 | 3-Indolylacrylic acid | 1.98 | 45.888 | 1.38×10-4 | 85 |
| 9 | Indole-3-pyruvic acid | 1.82 | 0.439 | 5.32×10-5 | 85 |
| 10 | Dihydrouracil | 1.79 | 15.319 | 6.22×10-7 | 100 |

图6 H2122-SR和H2122细胞的差异代谢通路富集分析Note: TCA cycle—tricarboxylic acid cycle.
Fig 6 Differential metabolic pathway enrichment analysis between H2122-SR and H2122 cells

图8 H2122-SR和H358-SR细胞与其亲本细胞的嘌呤代谢物热图分析Note: A. H2122 and H2122-SR cells. B. H358 and H358-SR cells. Red represents upregulation, and blue represents downregulation. R5P—ribose-5-phosphate; PRPP—phosphoribosyl pyrophosphate; IMP—inosine monophosphate; AMP—adenosine monophosphate; GMP—guanosine monophosphate.
Fig 8 Heatmap analysis of purine metabolites of H2122-SR and H358-SR cells with their parental cells
| 1 | BRAY F, LAVERSANNE M, SUNG H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2024, 74(3): 229-263. |
| 2 | RYAN M B, CORCORAN R B. Therapeutic strategies to target RAS-mutant cancers[J]. Nat Rev Clin Oncol, 2018, 15(11): 709-720. |
| 3 | MOORE A R, ROSENBERG S C, MCCORMICK F, et al. RAS-targeted therapies: is the undruggable drugged?[J]. Nat Rev Drug Discov, 2020, 19(8): 533-552. |
| 4 | JUDD J, ABDEL KARIM N, KHAN H, et al. Characterization of KRAS mutation subtypes in non-small cell lung cancer[J]. Mol Cancer Ther, 2021, 20(12): 2577-2584. |
| 5 | ZHU C X, GUAN X Q, ZHANG X N, et al. Targeting KRAS mutant cancers: from druggable therapy to drug resistance[J]. Mol Cancer, 2022, 21(1): 159. |
| 6 | SINGHAL A, LI B T, O'REILLY E M. Targeting KRAS in cancer[J]. Nat Med, 2024, 30(4): 969-983. |
| 7 | ZHANG J M, ZHANG J H, LIU Q, et al. Resistance looms for KRAS G12C inhibitors and rational tackling strategies[J]. Pharmacol Ther, 2022, 229: 108050. |
| 8 | YAEGER R, MEZZADRA R, SINOPOLI J, et al. Molecular characterization of acquired resistance to KRASG12C-EGFR inhibition in colorectal cancer[J]. Cancer Discov, 2023, 13(1): 41-55. |
| 9 | ZHAO Y L, MURCIANO-GOROFF Y R, XUE J Y, et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition[J]. Nature, 2021, 599(7886): 679-683. |
| 10 | 唐亚斌. 嘌呤代谢重编程ENPP1依赖机制在肺癌EGFR-TKI获得性耐药中的作用[D]. 上海: 上海交通大学, 2020. |
| TANG Y B. ENPP1-dependent purine metabolic reprogramming mechanism of EGFR-TKI acquired resistance in lung cancer[D]. Shanghai: Shanghai Jiao Tong University, 2020. | |
| 11 | RITCHIE M D, HOLZINGER E R, LI R W, et al. Methods of integrating data to uncover genotype-phenotype interactions[J]. Nat Rev Genet, 2015, 16(2): 85-97. |
| 12 | ORTMAYR K, DUBUIS S, ZAMPIERI M. Metabolic profiling of cancer cells reveals genome-wide crosstalk between transcriptional regulators and metabolism[J]. Nat Commun, 2019, 10(1): 1841. |
| 13 | VANDE VOORDE J, STEVEN R T, NAJUMUDEEN A K, et al. Metabolic profiling stratifies colorectal cancer and reveals adenosylhomocysteinase as a therapeutic target[J]. Nat Metab, 2023, 5(8): 1303-1318. |
| 14 | WANG Z Y, ZHU H Y, XIONG W. Advances in mass spectrometry-based multi-scale metabolomic methodologies and their applications in biological and clinical investigations[J]. Sci Bull, 2023, 68(19): 2268-2284. |
| 15 | HANAHAN D. Hallmarks of cancer: new dimensions[J]. Cancer Discov, 2022, 12(1): 31-46. |
| 16 | FAUBERT B, SOLMONSON A, DEBERARDINIS R J. Metabolic reprogramming and cancer progression[J]. Science, 2020, 368(6487): eaaw5473. |
| 17 | PANG Z Q, LU Y, ZHOU G Y, et al. MetaboAnalyst 6.0: towards a unified platform for metabolomics data processing, analysis and interpretation[J]. Nucleic Acids Res, 2024, 52(W1): W398-W406. |
| 18 | JIMÉNEZ-CARVELO A M, MARTÍN-TORRES S, ORTEGA-GAVILÁN F, et al. PLS-DA vs sparse PLS-DA in food traceability. A case study: authentication of avocado samples[J]. Talanta, 2021, 224: 121904. |
| 19 | SHAKEEL M, MAJEED M I, NAWAZ H, et al. Surface-enhanced Raman spectroscopy for the characterization of pellets of biofilm forming bacterial strains of Staphylococcus epidermidis[J]. Photodiagnosis Photodyn Ther, 2022, 40: 103145. |
| 20 | LIN Z Y, LI J W, ZHANG J, et al. Metabolic reprogramming driven by IGF2BP3 promotes acquired resistance to EGFR inhibitors in non-small cell lung cancer[J]. Cancer Res, 2023, 83(13): 2187-2207. |
| 21 | NIE M, CHEN N, PANG H H, et al. Targeting acetylcholine signaling modulates persistent drug tolerance in EGFR-mutant lung cancer and impedes tumor relapse[J]. J Clin Invest, 2022, 132(20): e160152. |
| 22 | SHI D D, SAVANI M R, ABDULLAH K G, et al. Emerging roles of nucleotide metabolism in cancer[J]. Trends Cancer, 2023, 9(8): 624-635. |
| 23 | MULLEN N J, SINGH P K. Nucleotide metabolism: a pan-cancer metabolic dependency[J]. Nat Rev Cancer, 2023, 23(5): 275-294. |
| 24 | SHUKLA S K, PUROHIT V, MEHLA K, et al. MUC1 and HIF-1α signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic cancer[J]. Cancer Cell, 2017, 32(3): 392. |
| 25 | HUANG Y T, CHAN S, CHEN S N, et al. Wnt/β-catenin signalling activates IMPDH2-mediated purine metabolism to facilitate oxaliplatin resistance by inhibiting caspase-dependent apoptosis in colorectal cancer[J]. J Transl Med, 2024, 22(1): 133. |
| 26 | ZHOU W H, YAO Y Y, SCOTT A J, et al. Purine metabolism regulates DNA repair and therapy resistance in glioblastoma[J]. Nat Commun, 2020, 11(1): 3811. |
| 27 | BEN-SAHRA I, HOXHAJ G, RICOULT S J H, et al. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle[J]. Science, 2016, 351(6274): 728-733. |
| 28 | HUANG F, NI M, CHALISHAZAR M D, et al. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers[J]. Cell Metab, 2018, 28(3): 369-382.e5. |
| 29 | TORRENCE M E, MACARTHUR M R, HOSIOS A M, et al. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals[J]. eLife, 2021, 10: e63326. |
| 30 | ALI E S, SAHU U, VILLA E, et al. ERK2 phosphorylates PFAS to mediate posttranslational control of de novo purine synthesis[J]. Mol Cell, 2020, 78(6): 1178-1191.e6. |
| 31 | XIAO Y, YU T J, XU Y, et al. Emerging therapies in cancer metabolism[J]. Cell Metab, 2023, 35(8): 1283-1303. |
| 32 | LABRIE M, BRUGGE J S, MILLS G B, et al. Therapy resistance: opportunities created by adaptive responses to targeted therapies in cancer[J]. Nat Rev Cancer, 2022, 22(6): 323-339. |
| 33 | WU Q, BA-ALAWI W, DEBLOIS G, et al. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer[J]. Nat Commun, 2020, 11(1): 4205. |
| 34 | LIU H, LYU H, JIANG G M, et al. ALKBH5-mediated m6A demethylation of GLUT4 mRNA promotes glycolysis and resistance to HER2-targeted therapy in breast cancer[J]. Cancer Res, 2022, 82(21): 3974-3986. |
| [1] | 黄昕, 刘家辉, 叶敬文, 钱文莉, 许万星, 王琳. 基于机器学习的小细胞肺癌代谢分子诊断模型的建立和临床应用[J]. 上海交通大学学报(医学版), 2025, 45(8): 1009-1016. |
| [2] | 张宇琴, 依力夏提·艾合买提, 王艳丽, 阳志, 黄建. MARCH9介导磷酸酶RPTPα的泛素化降解[J]. 上海交通大学学报(医学版), 2025, 45(8): 957-968. |
| [3] | 梁效宁, 石亭旺, 陈云丰. 小菌落变异株的致病机制及治疗研究进展[J]. 上海交通大学学报(医学版), 2025, 45(6): 784-791. |
| [4] | 张先洲, 杜凤麟, 吴雷, 任逸喆, 赵明娜, 娄加陶. OGT通过ERK信号通路促进非小细胞肺癌增殖的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(10): 1288-1297. |
| [5] | 朱鸣阳, 许元元, 任江浩, 黄嘉正, 李若楠, 谭强. 以磨玻璃结节为表现的肺腺癌亚肺叶切除研究综述[J]. 上海交通大学学报(医学版), 2024, 44(7): 922-927. |
| [6] | 王梦婷, 陈怡楠, 轩辕欣阳, 袁海花. 肺癌恶性胸腔积液来源肿瘤细胞的小鼠PDX模型构建及实验验证[J]. 上海交通大学学报(医学版), 2024, 44(4): 435-443. |
| [7] | 刘晨茜, 韩林, 杨轶, 周韩, 刘亚云, 盛德乔. GPR87通过激活RHO/ROCK通路促进非小细胞肺癌的侵袭和迁移[J]. 上海交通大学学报(医学版), 2024, 44(12): 1514-1525. |
| [8] | 黄华艳, 徐张闻笛, 夏立亮, 虞永峰, 陆舜. 表皮生长因子受体突变型晚期非小细胞肺癌免疫治疗的研究进展[J]. 上海交通大学学报(医学版), 2023, 43(5): 611-618. |
| [9] | 赵卓明, 刘振浩, 鲁曼曼, 张钰, 许林锋, 谢鹭. 基于TCR组库分析流程的非小细胞肺癌特征分析[J]. 上海交通大学学报(医学版), 2023, 43(12): 1520-1528. |
| [10] | 赵富茂, 彭玫, 彭晓露, 舒韦韦, 彭丽. 鲍曼不动杆菌在环境美罗培南浓度变化时耐药性的改变及其机制[J]. 上海交通大学学报(医学版), 2023, 43(11): 1396-1407. |
| [11] | 邹菁华, 宫淼淼, 沈瑛. KRAS4AG12C和KRAS4BG12C对人肺上皮细胞生长和运动的作用差异及机制研究[J]. 上海交通大学学报(医学版), 2022, 42(8): 1016-1023. |
| [12] | 赵珂珂, 蒋蓓蓓, 张璐, 王凌云, 张亚平, 解学乾. 超低剂量平扫CT深度学习图像重建评价肺部病灶的可行性[J]. 上海交通大学学报(医学版), 2022, 42(8): 1062-1069. |
| [13] | 廖雅慧, 刘丽云, 朱泓睿, 林厚文, 严继舟, 孙凡. 海绵来源的smenospongine通过抑制非小细胞肺癌细胞中的EGFR-Akt-ABCG2信号通路抑制顺铂耐药[J]. 上海交通大学学报(医学版), 2022, 42(8): 997-1007. |
| [14] | 刘子杨, 王小文, 陈力. lncRNA GK-IT1通过调控醛缩酶A影响非小细胞肺癌细胞的恶性进展[J]. 上海交通大学学报(医学版), 2022, 42(5): 591-601. |
| [15] | 陆文清, 孟周文理, 虞永峰, 陆舜. 非小细胞肺癌第三代表皮生长因子受体-酪氨酸激酶抑制剂的耐药机制及治疗策略[J]. 上海交通大学学报(医学版), 2022, 42(4): 535-544. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||