
上海交通大学学报(医学版) ›› 2025, Vol. 45 ›› Issue (4): 387-403.doi: 10.3969/j.issn.1674-8115.2025.04.001
• 论著 · 基础研究 • 下一篇
李林颖1, 蔡晓东2, 童冉2, 杨晨2, 王志明2, 贺潇宇2, 马子越2, 张丰2( ), 李令杰2(
), 李令杰2( ), 周君梅1(
), 周君梅1( )
)
收稿日期:2024-09-02
接受日期:2024-11-11
出版日期:2025-04-28
发布日期:2025-04-28
通讯作者:
周君梅,研究员,博士;电子信箱:zhou_junmei@qq.com作者简介:李林颖(1998—),女,硕士生;电子信箱:lin-ying@sjtu.edu.cn。
基金资助:
LI Linying1, CAI Xiaodong2, TONG Ran2, YANG Chen2, WANG Zhiming2, HE Xiaoyu2, MA Ziyue2, ZHANG Feng2( ), LI Lingjie2(
), LI Lingjie2( ), ZHOU Junmei1(
), ZHOU Junmei1( )
)
Received:2024-09-02
Accepted:2024-11-11
Online:2025-04-28
Published:2025-04-28
Contact:
ZHOU Junmei, E-mail: zhou_junmei@qq.comSupported by:摘要:
目的·利用人胚胎干细胞(human embryonic stem cell,hESC)体外分化模型和高通量多组学测序技术研究hESC分化成神经前体细胞(neural progenitor cell,NPC)过程中转录组和染色质可及性的变化情况。方法·首先在体外利用拟胚体形成法诱导hESC分化成NPC,并收集这2个阶段的细胞;通过反转录-实时荧光定量PCR(reverse transcription-quantitative real-time PCR,RT-qPCR)和免疫荧光染色(immunofluorescence,IF)鉴定细胞表型。应用转录组测序(transcriptome sequencing,RNA-seq)检测并分析hESC和NPC的差异表达基因(differentially expressed gene,DEG)。应用染色质可及性测序(assay for transposase-accessible chromatin with high throughput sequencing,ATAC-seq)技术获取hESC和NPC的染色质可及性变化情况,并对差异的染色质开放区域进行基序富集分析以发现具有潜在调控作用的转录因子。最后对RNA-seq和ATAC-seq多组学数据进行联合分析,并构建蛋白质互作(protein-protein interaction,PPI)网络,寻找体外神经早期分化过程中的关键基因和调控通路。结果·RT-qPCR与IF均显示多能性标志物(NANOG、POU5F1)在hESC阶段表达量高而在NPC阶段表达量明显降低;同时神经早期分化标志物(PAX6、SOX1、NES)在hESC阶段基本不表达而在NPC阶段表达量显著升高。RNA-seq分析结果显示,与hESC阶段相比,NPC阶段中有5 597个基因的表达水平呈现上调,而3 654个基因的表达水平下降,基因功能富集分析显示NPC阶段上调的基因富集至神经发育相关的功能。ATAC-seq分析结果显示,共27 491个基因组区域在hESC向NPC分化过程中染色质可及性发生了显著改变,其中有12 381个区域染色质可及性增强,15 110个区域染色质可及性减弱;基序富集分析揭示DLX1、LHX2等转录因子基因可能在hESC向NPC分化过程中发挥重要作用。RNA-seq和ATAC-seq的多组学数据联合分析结果显示,在NPC阶段高表达的重叠基因主要富集在轴突导向、前脑发育、神经元迁移等。神经分化后CTNND2、LHX2基因表达水平升高,且相关基因区域染色质可及性也增加。PPI网络分析发现,PRKACA、CDH2、ERBB4等是下游候选基因。结论·利用hESC体外分化模型结合高通量多组学测序技术可用于揭示hESC向NPC分化过程中的转录组及染色质可及性的变化规律;该过程中轴突导向、前脑发育、神经元迁移等通路的相关基因表达水平升高,染色质可及性增强。
中图分类号:
李林颖, 蔡晓东, 童冉, 杨晨, 王志明, 贺潇宇, 马子越, 张丰, 李令杰, 周君梅. 人胚胎干细胞向神经前体细胞分化的转录组与染色质可及性变化分析研究[J]. 上海交通大学学报(医学版), 2025, 45(4): 387-403.
LI Linying, CAI Xiaodong, TONG Ran, YANG Chen, WANG Zhiming, HE Xiaoyu, MA Ziyue, ZHANG Feng, LI Lingjie, ZHOU Junmei. Analysis of transcriptome and chromatin accessibility changes during the differentiation of human embryonic stem cells into neural progenitor cells[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(4): 387-403.
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| GAPDH | CTGAGAACGGGAAGCTTGT | GGGTGCTAAGCAGTTGGT |
| NANOG | GAATGAAATCTAAGAGGTGGCA | CCTGGTGGTAGGAAGAGTAAAGG |
| POU5F1 | ACATCAAAGCTCTGCAGAAAGAACT | CTGAATACCTTCCCAAATAGAACCC |
| SOX1 | GGAATGGGAGGACAGGATTT | ACTTTTATTTCTCGGCCCGT |
| PAX6 | GCCTATGCAACCCCCAGT | TCACTTCCGGGAACTTGAAC |
| NES | TGCGGGCTACTGAAAAGTTC | AGGCTGAGGGACATCTTGAG |
表1 RT-qPCR引物序列
Tab 1 Primer sequences for RT-qPCR
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| GAPDH | CTGAGAACGGGAAGCTTGT | GGGTGCTAAGCAGTTGGT |
| NANOG | GAATGAAATCTAAGAGGTGGCA | CCTGGTGGTAGGAAGAGTAAAGG |
| POU5F1 | ACATCAAAGCTCTGCAGAAAGAACT | CTGAATACCTTCCCAAATAGAACCC |
| SOX1 | GGAATGGGAGGACAGGATTT | ACTTTTATTTCTCGGCCCGT |
| PAX6 | GCCTATGCAACCCCCAGT | TCACTTCCGGGAACTTGAAC |
| NES | TGCGGGCTACTGAAAAGTTC | AGGCTGAGGGACATCTTGAG |
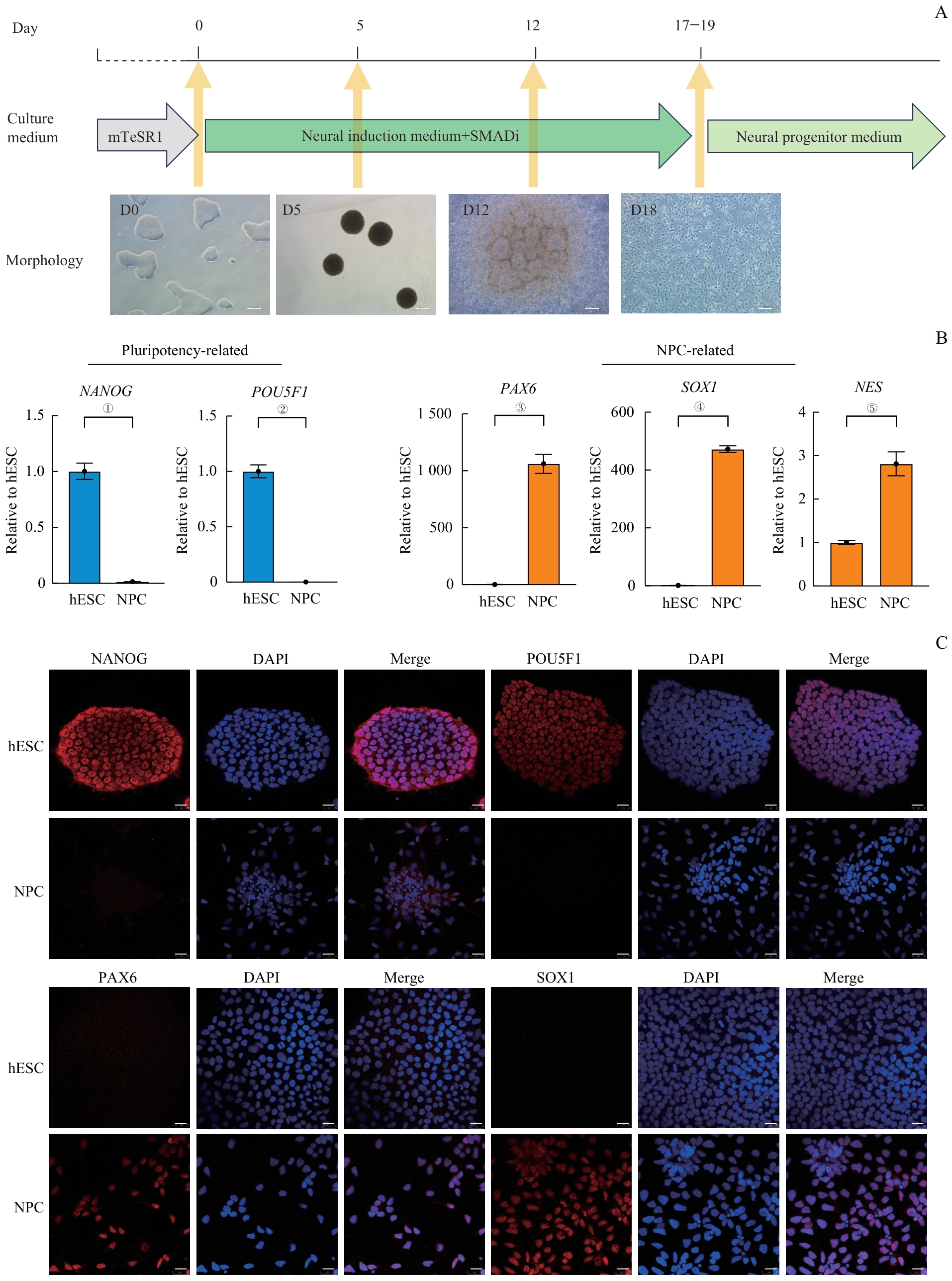
图1 利用双重SMAD抑制策略在体外将hESC分化成NPCNote:A. Schematic diagram of the NPC culture system and morphology of hESC-induced differentiation into NPCs. SMADi—SMAD inhibition. Scale bar=200 μm. B. The gene expression of typical genes related to pluripotency and NPCs during the induction process detected by RT-qPCR. ①P=0.002, ②P=0.001, ③P=0.002, ④P<0.001, ⑤P=0.006. NPCs were the cells induced for 27 d. C. Expression of hESC markers (NANOG and POU5F1) and NPC markers (PAX6 and SOX1), detected by immunofluorescence staining. NPCs were the cells induced for 27 d. Scale bar=25 μm.
Fig 1 Dual SMAD inhibition strategy for hESC differentiation into NPC in vitro
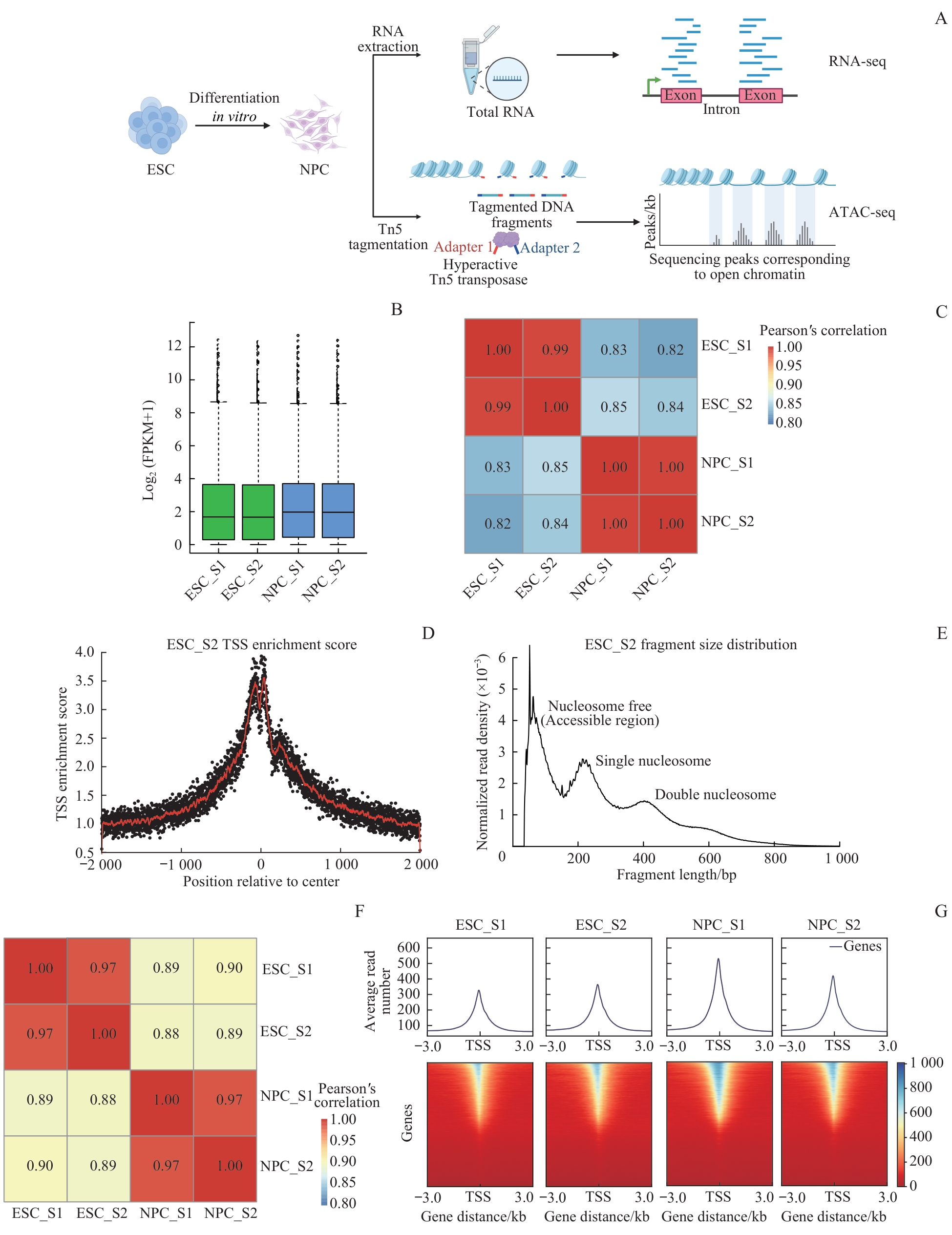
图2 体外神经早期分化过程中RNA-seq和ATAC-seq质控分析Note: A. Experimental flow chart. Cells were collected at the ESC stage and NPC stage; nuclei were extracted for standard ATAC-seq library preparation, while total RNA was extracted for RNA-seq library preparation. B. Box plot of gene expression levels. C. Pearson correlation analysis was performed on the expression levels between different samples. D. Transcription start site (TSS) enrichment scores of ATAC-seq data. E. Fragment size distribution of ATAC-seq data. F. Pearson correlation analysis of ATAC-seq data between different samples. G. Gene TSS enrichment heat map showing the reads from ATAC-seq data in the TSS±3.0 kb. The blue-to-red scale represents the transition from high to low in the reads in the specified area.
Fig 2 Quality control analysis of RNA-seq and ATAC-seq datasets obtained from the process of early neural differentiation in vitro
| Index | Sample | |||
|---|---|---|---|---|
| ESC_S1 | ESC_S2 | NPC_S1 | NPC_S2 | |
| Total raw read pairs/n | 45 792 974 | 53 251 136 | 50 419 620 | 52 973 024 |
| Total raw bases/n | 6 914 739 074 | 8 040 921 536 | 7 613 362 620 | 7 998 926 624 |
| Mapped reads/n | 44 506 927 | 51 742 966 | 48 948 795 | 51 574 332 |
| Mapped ratio/% | 97.95 | 97.92 | 97.87 | 98.13 |
| High-quality mapped reads/n | 43 139 843 | 50 148 322 | 47 576 740 | 50 093 517 |
| High-quality mapped ratio/% | 94.94 | 94.90 | 95.13 | 95.31 |
表2 在神经诱导过程中RNA-seq文库所获得的原始读段数和比对读段数
Tab 2 Number of raw reads and mapped reads obtained from RNA-seq libraries during neural induction
| Index | Sample | |||
|---|---|---|---|---|
| ESC_S1 | ESC_S2 | NPC_S1 | NPC_S2 | |
| Total raw read pairs/n | 45 792 974 | 53 251 136 | 50 419 620 | 52 973 024 |
| Total raw bases/n | 6 914 739 074 | 8 040 921 536 | 7 613 362 620 | 7 998 926 624 |
| Mapped reads/n | 44 506 927 | 51 742 966 | 48 948 795 | 51 574 332 |
| Mapped ratio/% | 97.95 | 97.92 | 97.87 | 98.13 |
| High-quality mapped reads/n | 43 139 843 | 50 148 322 | 47 576 740 | 50 093 517 |
| High-quality mapped ratio/% | 94.94 | 94.90 | 95.13 | 95.31 |
| Index | Sample | |||
|---|---|---|---|---|
| ESC_S1 | ESC_S2 | NPC_S1 | NPC_S2 | |
| Total raw read pairs/n | 63 713 180 | 32 001 236 | 88 523 604 | 61 924 682 |
| Total raw bases/n | 9 620 690 180 | 4 832 186 636 | 13 367 000 000 | 9 350 626 982 |
| Mapped read pairs/n | 61 240 240 | 30 684 142 | 84 944 932 | 59 439 388 |
| Mapped ratio/% | 98.36 | 98.52 | 98.24 | 97.98 |
| High-quality mapped read pairs/n | 54 624 252 | 27 152 448 | 75 936 202 | 53 201 562 |
| High-quality mapped ratio/% | 87.73 | 87.18 | 87.82 | 87.70 |
| Percent duplication/% | 23.70 | 20.59 | 22.18 | 29.05 |
| Non-duplicate read pairs/n | 41 677 046 | 21 562 952 | 59 092 184 | 37 745 230 |
表3 在神经诱导过程中ATAC-seq文库所获得的原始读段数和比对读段数
Tab 3 Number of raw reads and mapped reads obtained by ATAC-seq libraries during neural induction
| Index | Sample | |||
|---|---|---|---|---|
| ESC_S1 | ESC_S2 | NPC_S1 | NPC_S2 | |
| Total raw read pairs/n | 63 713 180 | 32 001 236 | 88 523 604 | 61 924 682 |
| Total raw bases/n | 9 620 690 180 | 4 832 186 636 | 13 367 000 000 | 9 350 626 982 |
| Mapped read pairs/n | 61 240 240 | 30 684 142 | 84 944 932 | 59 439 388 |
| Mapped ratio/% | 98.36 | 98.52 | 98.24 | 97.98 |
| High-quality mapped read pairs/n | 54 624 252 | 27 152 448 | 75 936 202 | 53 201 562 |
| High-quality mapped ratio/% | 87.73 | 87.18 | 87.82 | 87.70 |
| Percent duplication/% | 23.70 | 20.59 | 22.18 | 29.05 |
| Non-duplicate read pairs/n | 41 677 046 | 21 562 952 | 59 092 184 | 37 745 230 |
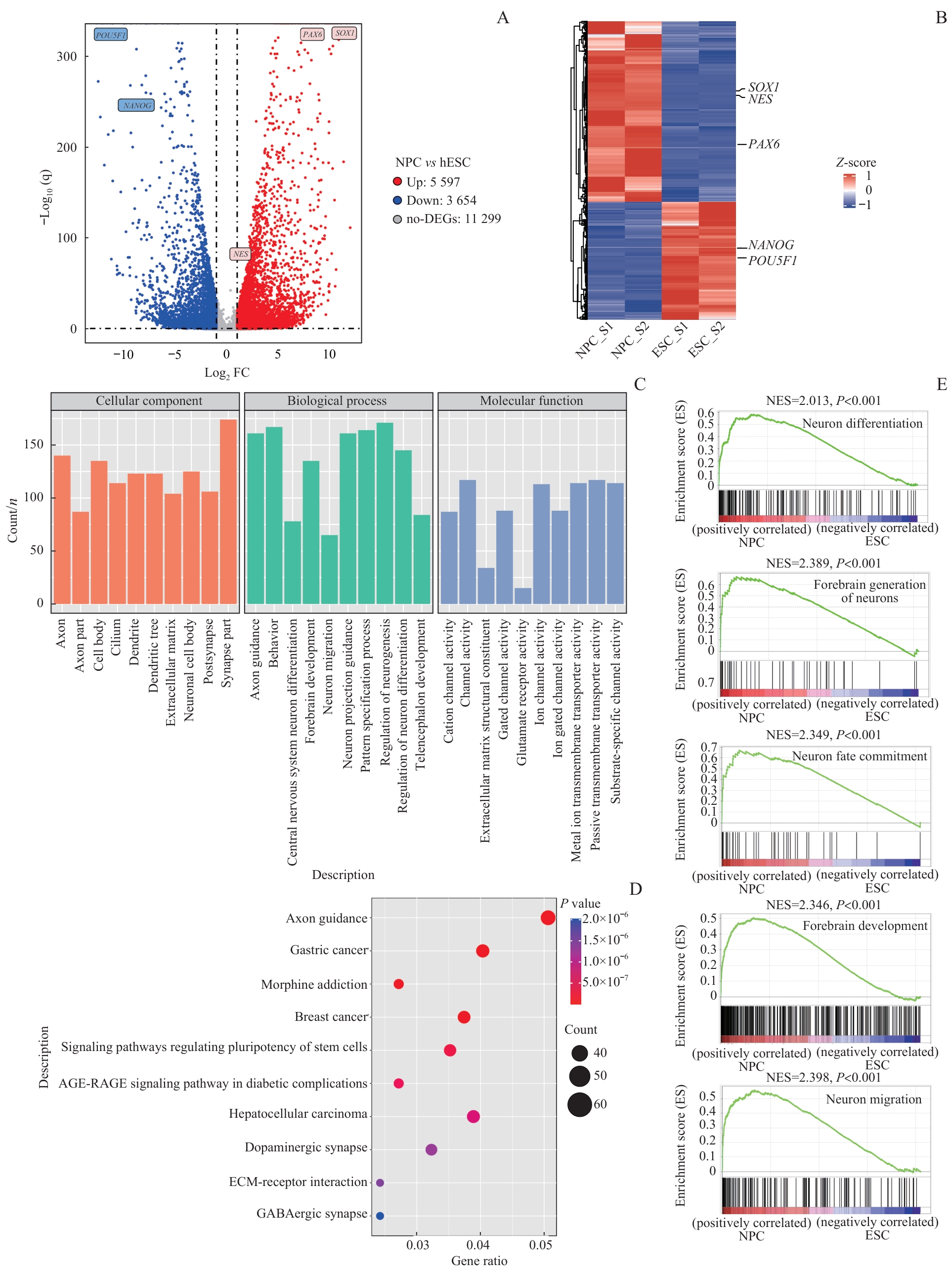
图3 NPC分化过程中转录组的分析Note: A. Volcano plot shows the distribution of DEGs between the two distinct developmental stages (ESC and NPC). DEGs were defined as |log2FC|>1 and P<0.05. B. Gene expression levels were clustered across all samples. Different colors indicate the value of each gene expression level normalized with Z-scores. C. GO analysis was performed on the DEGs upregulated at the NPC stage. D. KEGG analysis of the DEGs upregulated at NPC stage. E. GSEA analysis identifies pathways upregulated during NPC differentiation.
Fig 3 Transcriptome analysis of NPC differentiation
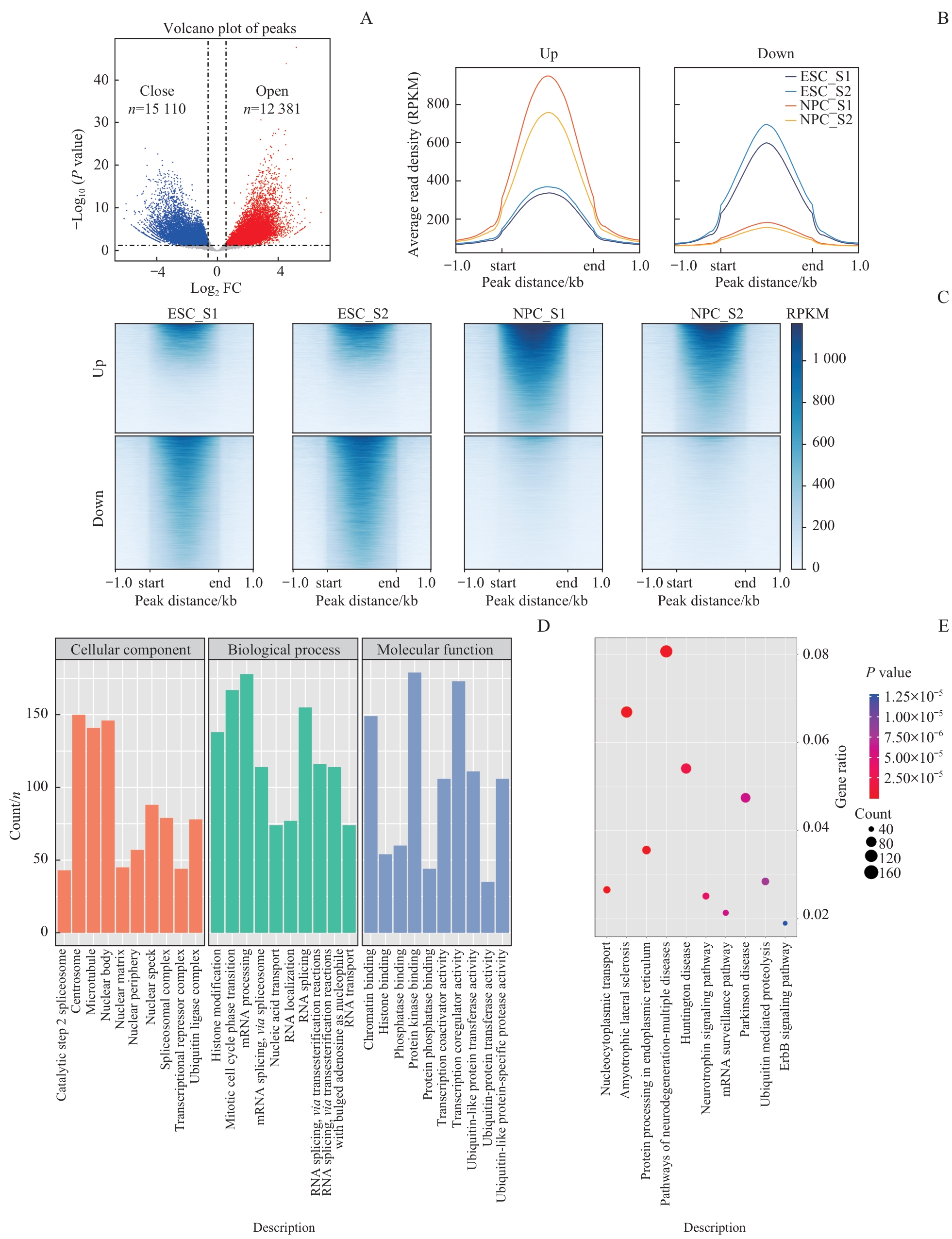
图4 NPC分化过程中染色质可及性分析Note: A. Volcano plot shows the peak signals of differentially open chromatin obtained from the ATAC-seq dataset. Differential open areas were defined as |log2FC| >0.585 and P<0.05. B. The differential region enrichment plot shows the average peak signal between different samples in the differentially upregulated region (left) and the differentially downregulated region (right) at the NPC stage. C. Differential region enrichment heatmap reveals peak signals within genomic region. D. GO analysis of genes associated with differentially open chromatin regions at the NPC stage. E. KEGG analysis of genes associated with differentially open chromatin regions at the NPC stage.
Fig 4 Chromatin accessibility analysis during NPC differentiation process
| Motif | E value | Motif ID | Motif | E value | Motif ID |
|---|---|---|---|---|---|
 | 2.38×10-111 | DLX1 | 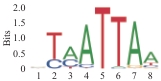 | 1.05×10-100 | NKX6.1 |
 | 6.26×10-109 | SHOX |  | 2.79×10-91 | LHX2 |
 | 6.18×10-101 | MSX1 |
表4 对在NPC差异上调的开放染色质信号进行基序富集分析
Tab 4 Motif enrichment analysis of open chromatin signals that were differentially upregulated at the NPC stage
| Motif | E value | Motif ID | Motif | E value | Motif ID |
|---|---|---|---|---|---|
 | 2.38×10-111 | DLX1 |  | 1.05×10-100 | NKX6.1 |
 | 6.26×10-109 | SHOX |  | 2.79×10-91 | LHX2 |
 | 6.18×10-101 | MSX1 |
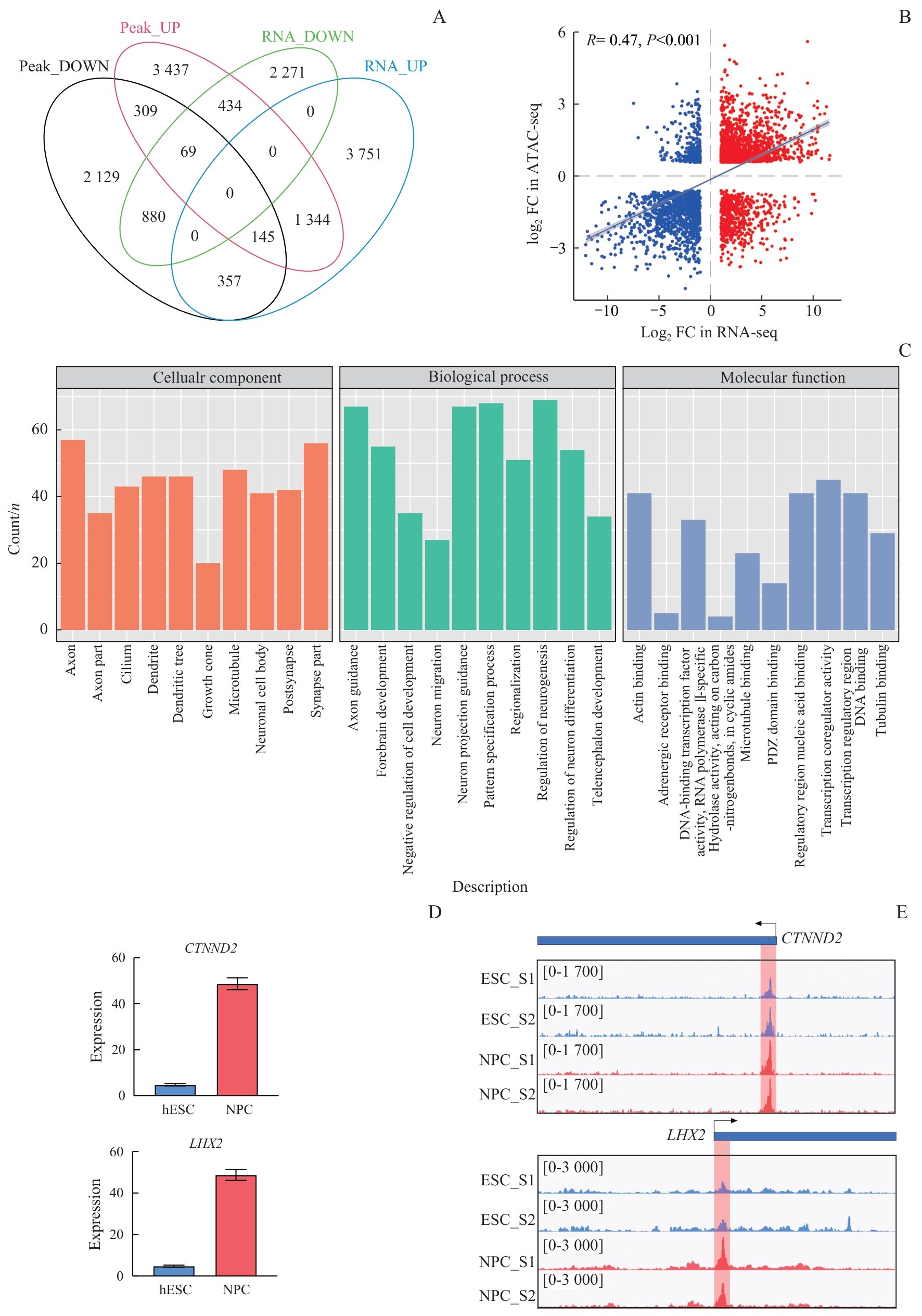
图5 整合RNA-seq和ATAC-seq数据分析参与NPC分化的重要基因Note: A. Venn diagram showing the overlap between DEGs in the NPC cell population and related genes predicted from differentially open chromatin regions. In the ATAC-seq dataset, genes were annotated to regions with ±1 kb of the promoter. B. Scatter plot showing the correlation between the expression levels of the overlapping genes (A) obtained in RNA-seq and the associated peaks obtained from ATAC-seq. C. GO analysis of 1 344 upregulated genes. D. Expression levels of CTNND2 and LHX2 in RNA-seq during NPC differentiation. E. The Integrative Genomics Viewer showing ATAC-seq signals at the transcription start sites of CTNND2 and LHX2 loci during NPC differentiation.
Fig 5 Integration of RNA-seq and ATAC-seq data to analyze important genes involved in NPC differentiation
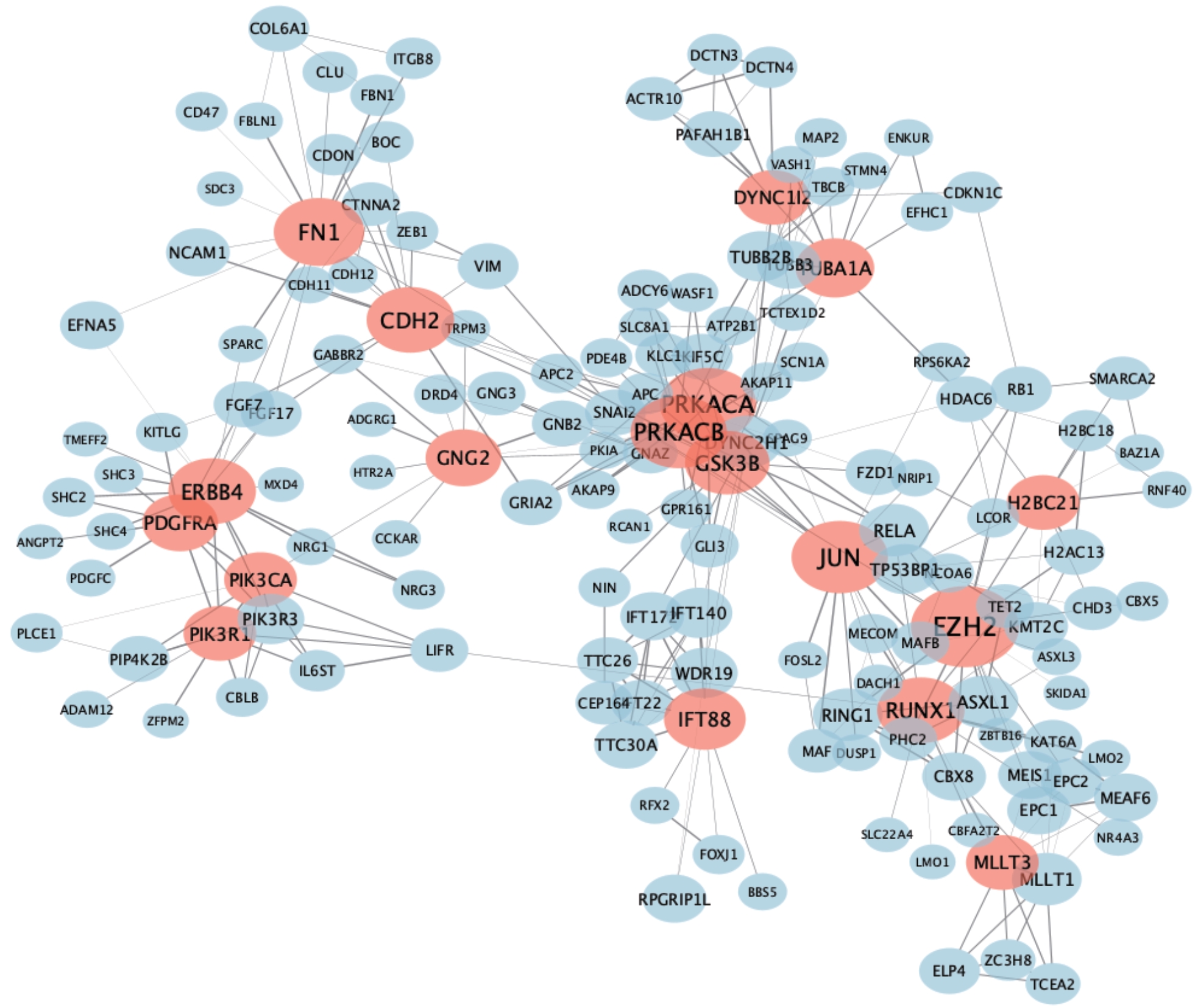
图6 PPI网络分析揭示下游候选中心基因Note: The thickness of the edge in the network diagram represents the interaction strength between the two proteins, and the size of the node represents the degree. The red circles represent the protein with degree≥10, and the others represent proteins that interact with the red proteins.
Fig 6 Candidate hub downstream genes detected by PPI network analysis
| Gene | Degree |
|---|---|
| EZH2 | 22 |
| PRKACA | 18 |
| PRKACB | 18 |
| JUN | 18 |
| FN1 | 16 |
| CDH2 | 15 |
| ERBB4 | 15 |
| RUNX1 | 15 |
| GSK3B | 14 |
| IFT88 | 13 |
| TUBA1A | 12 |
| PDGFRA | 11 |
| GNG2 | 11 |
| DYNC1I2 | 10 |
| H2BC21 | 10 |
| PIK3CA | 10 |
| PIK3R1 | 10 |
| MLLT3 | 10 |
表5 PPI网络中度≥10的下游候选中心基因
Tab 5 Candidate downstream hub genes with the degrees≥10 in the PPI network
| Gene | Degree |
|---|---|
| EZH2 | 22 |
| PRKACA | 18 |
| PRKACB | 18 |
| JUN | 18 |
| FN1 | 16 |
| CDH2 | 15 |
| ERBB4 | 15 |
| RUNX1 | 15 |
| GSK3B | 14 |
| IFT88 | 13 |
| TUBA1A | 12 |
| PDGFRA | 11 |
| GNG2 | 11 |
| DYNC1I2 | 10 |
| H2BC21 | 10 |
| PIK3CA | 10 |
| PIK3R1 | 10 |
| MLLT3 | 10 |
| 1 | UCHIDA N, BUCK D W, HE D, et al. Direct isolation of human central nervous system stem cells[J]. Proc Natl Acad Sci USA, 2000, 97(26): 14720-14725. |
| 2 | NOCTOR S C, FLINT A C, WEISSMAN T A, et al. Neurons derived from radial glial cells establish radial units in neocortex[J]. Nature, 2001, 409(6821): 714-720. |
| 3 | KRIEGSTEIN A, ALVAREZ-BUYLLA A. The glial nature of embryonic and adult neural stem cells[J]. Annu Rev Neurosci, 2009, 32: 149-184. |
| 4 | MARCHETTO M C, BELINSON H, TIAN Y, et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals[J]. Mol Psychiatry, 2017, 22(6): 820-835. |
| 5 | HOWELL B W, SMITH K M. Synaptic structural protein dysfunction leads to altered excitation inhibition ratios in models of autism spectrum disorder[J]. Pharmacol Res, 2019, 139: 207-214. |
| 6 | BOZZI Y, PROVENZANO G, CASAROSA S. Neurobiological bases of autism-epilepsy comorbidity: a focus on excitation/inhibition imbalance[J]. Eur J Neurosci, 2018, 47(6): 534-548. |
| 7 | OLIVEIRA B, MITJANS M, NITSCHE M A, et al. Excitation-inhibition dysbalance as predictor of autistic phenotypes[J]. J Psychiatr Res, 2018, 104: 96-99. |
| 8 | MOOSAVI A, MOTEVALIZADEH ARDEKANI A. Role of epigenetics in biology and human diseases[J]. Iran Biomed J, 2016, 20(5): 246-258. |
| 9 | FITZ-JAMES M H, CAVALLI G. Molecular mechanisms of transgenerational epigenetic inheritance[J]. Nat Rev Genet, 2022, 23(6): 325-341. |
| 10 | ALLIS C D, JENUWEIN T. The molecular hallmarks of epigenetic control[J]. Nat Rev Genet, 2016, 17(8): 487-500. |
| 11 | DAI S K, LIU P P, LI X, et al. Dynamic profiling and functional interpretation of histone lysine crotonylation and lactylation during neural development[J]. Development, 2022, 149(14): dev200049. |
| 12 | SUN T Y, XU Y Y, XIANG Y, et al. Crosstalk between RNA m6A and DNA methylation regulates transposable element chromatin activation and cell fate in human pluripotent stem cells[J]. Nat Genet, 2023, 55(8): 1324-1335. |
| 13 | WANG C F, YANG J W, ZHUANG Z H, et al. Activity-dependent feedback regulation of thalamocortical axon development by Lhx2 in cortical layer 4 neurons[J]. Cereb Cortex, 2023, 33(5): 1693-1707. |
| 14 | TSENG C J, MCDOUGLE C J, HOOKER J M, et al. Epigenetics of autism spectrum disorder: histone deacetylases[J]. Biol Psychiatry, 2022, 91(11): 922-933. |
| 15 | CONBOY K, HENSHALL D C, BRENNAN G P. Epigenetic principles underlying epileptogenesis and epilepsy syndromes[J]. Neurobiol Dis, 2021, 148: 105179. |
| 16 | KLEMM S L, SHIPONY Z, GREENLEAF W J. Chromatin accessibility and the regulatory epigenome[J]. Nat Rev Genet, 2019, 20(4): 207-220. |
| 17 | SONG L Y, CRAWFORD G E. [J]. Cold Spring Harb Protoc, 2010, 2010(2): pdb.prot5384. |
| 18 | BUENROSTRO J D, GIRESI P G, ZABA L C, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position[J]. Nat Methods, 2013, 10(12): 1213-1218. |
| 19 | MARKENSCOFF-PAPADIMITRIOU E, WHALEN S, PRZYTYCKI P, et al. A chromatin accessibility atlas of the developing human telencephalon[J]. Cell, 2020, 182(3): 754-769.e18. |
| 20 | LU C Y, GARIPLER G, DAI C, et al. Essential transcription factors for induced neuron differentiation[J]. Nat Commun, 2023, 14(1): 8362. |
| 21 | THOMSON J A, ITSKOVITZ-ELDOR J, SHAPIRO S S, et al. Embryonic stem cell lines derived from human blastocysts[J]. Science, 1998, 282(5391): 1145-1147. |
| 22 | TAKAHASHI K, TANABE K, OHNUKI M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors[J]. Cell, 2007, 131(5): 861-872. |
| 23 | YU J Y, VODYANIK M A, SMUGA-OTTO K, et al. Induced pluripotent stem cell lines derived from human somatic cells[J]. Science, 2007, 318(5858): 1917-1920. |
| 24 | CHAMBERS S M, FASANO C A, PAPAPETROU E P, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling[J]. Nat Biotechnol, 2009, 27(3): 275-280. |
| 25 | KAWASAKI H, MIZUSEKI K, NISHIKAWA S, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity[J]. Neuron, 2000, 28(1): 31-40. |
| 26 | LEE H, SHAMY G A, ELKABETZ Y, et al. Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons[J]. Stem Cells, 2007, 25(8): 1931-1939. |
| 27 | ZHANG S C, WERNIG M, DUNCAN I D, et al. In vitro differentiation of transplantable neural precursors from human embryonic stem cells[J]. Nat Biotechnol, 2001, 19(12): 1129-1133. |
| 28 | TCHIEU J, ZIMMER B, FATTAHI F, et al. A modular platform for differentiation of human PSCs into all major ectodermal lineages[J]. Cell Stem Cell, 2017, 21(3): 399-410.e7. |
| 29 | MARTIN M. Cutadapt removes adapter sequences from high-throughput sequencing reads[J]. EMBnet J, 2011, 17(1): 10-12. |
| 30 | BOLGER A M, LOHSE M, USADEL B. Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30(15): 2114-2120. |
| 31 | PATEL R K, JAIN M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data[J]. PLoS One, 2012, 7(2): e30619. |
| 32 | KIM D, LANGMEAD B, SALZBERG S L. HISAT: a fast spliced aligner with low memory requirements[J]. Nat Methods, 2015, 12(4): 357-360. |
| 33 | LIAO Y, SMYTH G K, SHI W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features[J]. Bioinformatics, 2014, 30(7): 923-930. |
| 34 | LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. |
| 35 | YU G C, WANG L G, HAN Y Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16(5): 284-287. |
| 36 | LANGMEAD B, SALZBERG S L. Fast gapped-read alignment with bowtie 2[J]. Nat Methods, 2012, 9(4): 357-359. |
| 37 | ZHANG Y, LIU T, MEYER C A, et al. Model-based analysis of ChIP-seq (MACS)[J]. Genome Biol, 2008, 9(9): R137. |
| 38 | BAILEY T L, BODEN M, BUSKE F A, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| 39 | ROSS-INNES C S, STARK R, TESCHENDORFF A E, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer[J]. Nature, 2012, 481(7381): 389-393. |
| 40 | WATANABE K, UENO M, KAMIYA D, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells[J]. Nat Biotechnol, 2007, 25(6): 681-686. |
| 41 | ITSKOVITZ-ELDOR J, SCHULDINER M, KARSENTI D, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers[J]. Mol Med, 2000, 6(2): 88-95. |
| 42 | SZKLARCZYK D, KIRSCH R, KOUTROULI M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest[J]. Nucleic Acids Res, 2023, 51(D1): D638-D646. |
| 43 | JIA E T, PAN M, LIU Z Y, et al. Transcriptomic profiling of differentially expressed genes and related pathways in different brain regions in Parkinson's disease[J]. Neurosci Lett, 2020, 732: 135074. |
| 44 | ARAYA C, HÄKKINEN H M, CARCAMO L, et al. Cdh2 coordinates Myosin-Ⅱ dependent internalisation of the zebrafish neural plate[J]. Sci Rep, 2019, 9(1): 1835. |
| 45 | BARETTINO C, BALLESTEROS-GONZALEZ Á, AYLÓN A, et al. Developmental disruption of Erbb4 in Pet1+ neurons impairs serotonergic sub-system connectivity and memory formation[J]. Front Cell Dev Biol, 2021, 9: 770458. |
| 46 | TUNCAY I O, PARMALEE N L, KHALIL R, et al. Analysis of recent shared ancestry in a familial cohort identifies coding and noncoding autism spectrum disorder variants[J]. NPJ Genom Med, 2022, 7(1): 13. |
| 47 | ASADOLLAHI R, ONEDA B, JOSET P, et al. The clinical significance of small copy number variants in neurodevelopmental disorders[J]. J Med Genet, 2014, 51(10): 677-688. |
| 48 | CHEN Z N, LI X J, CUI X Z, et al. Association of CTNND2 gene polymorphism with schizophrenia: two-sample case-control study in Chinese Han population[J]. Int J Psychiatry Med, 2023, 58(5): 433-448. |
| 49 | WANG L Y, XU M, WANG Y, et al. Melatonin improves synapse development by PI3K/Akt signaling in a mouse model of autism spectrum disorder[J]. Neural Regen Res, 2024, 19(7): 1618-1624. |
| 50 | VAZ R, EDWARDS S, DUEÑAS-REY A, et al. Loss of ctnnd2b affects neuronal differentiation and behavior in zebrafish[J]. Front Neurosci, 2023, 17: 1205653. |
| 51 | SCHMID C M, GREGOR A, COSTAIN G, et al. LHX2 haploinsufficiency causes a variable neurodevelopmental disorder[J]. Genet Med, 2023, 25(7): 100839. |
| 52 | WANG Y N, KHANDELWAL N, LIU S Q, et al. KDM6B cooperates with Tau and regulates synaptic plasticity and cognition via inducing VGLUT1/2[J]. Mol Psychiatry, 2022, 27(12): 5213-5226. |
| 53 | LI W, SHEN W C, ZHANG B, et al. Long non-coding RNA LncKdm2b regulates cortical neuronal differentiation by cis-activating Kdm2b[J]. Protein Cell, 2020, 11(3): 161-186. |
| 54 | CHEN D H, MCMANUS C E, RADMANESH B, et al. Temporal inhibition of chromatin looping and enhancer accessibility during neuronal remodeling[J]. Nat Commun, 2021, 12(1): 6366. |
| 55 | RAHMAN S, DONG P F, APONTES P, et al. Lineage specific 3D genome structure in the adult human brain and neurodevelopmental changes in the chromatin interactome[J]. Nucleic Acids Res, 2023, 51(20): 11142-11161. |
| 56 | CORCES M R, SHCHERBINA A, KUNDU S, et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer's and Parkinson's diseases[J]. Nat Genet, 2020, 52(11): 1158-1168. |
| 57 | PAGNI S, MILLS J D, FRANKISH A, et al. Non-coding regulatory elements: potential roles in disease and the case of epilepsy[J]. Neuropathol Appl Neurobiol, 2022, 48(3): e12775. |
| [1] | 霍晓燕, 罗小梅, 叶贤涛, 孙昱, 余永国, 梁黎黎, 范燕洁. MMAA基因非编码区变异叠加单亲二体所致甲基丙二酸血症的多组学分析[J]. 上海交通大学学报(医学版), 2025, 45(6): 800-806. |
| [2] | 禹志远, 董海平, 高楠, 马柯. 背根神经节吗啡耐受核心基因筛选与机制研究:加权基因共表达网络分析和机器学习的转录组学整合策略[J]. 上海交通大学学报(医学版), 2025, 45(10): 1308-1319. |
| [3] | 梁乐斌, 陈慧芳, 赖淑静, 顾靓, 苏冰. 基于空间ATAC-seq技术的Apcmin/+小鼠结肠肿瘤表观特征分析[J]. 上海交通大学学报(医学版), 2025, 45(10): 1261-1270. |
| [4] | 刘思雨, 张磊. 七氟烷抑制新生小鼠前额叶皮质神经祖细胞向神经元分化发育[J]. 上海交通大学学报(医学版), 2023, 43(9): 1115-1130. |
| [5] | 周月, 程晨, 郑恩霖, 孟卓, 王鉴, 王青洁, 何勇宁, 孙锟. 利用Crispr/Cas9基因编辑系统在人胚胎干细胞中探索ELABELA的潜在新受体[J]. 上海交通大学学报(医学版), 2022, 42(9): 1258-1264. |
| [6] | 赵冰楠, 马双羽, 胥春龙, 王琼. SMAD2和SMAD3在人胚胎干细胞分化过程中的不同调控作用[J]. 上海交通大学学报(医学版), 2022, 42(4): 443-454. |
| [7] | 孙潇智, 李爽, 金颖, 廖兵. 酶消化细胞团块法对人胚胎干细胞中OCT4与SOX2蛋白水平的影响[J]. 上海交通大学学报(医学版), 2021, 41(4): 413-420. |
| [8] | 丁 蕾1, 2,高彩霞1, 2,刘兆远1, 2,陈 磊1, 2. 少量细胞输入的自制转录组测序文库构建试剂评测[J]. 上海交通大学学报(医学版), 2020, 40(4): 472-. |
| [9] | 朱超南,陈沁雯,辛晨歌,李慧. 人胚胎干细胞多基因同时抑制系统的建立[J]. 上海交通大学学报(医学版), 2019, 39(5): 478-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||