
上海交通大学学报(医学版) ›› 2025, Vol. 45 ›› Issue (9): 1171-1182.doi: 10.3969/j.issn.1674-8115.2025.09.009
朱子俊1, 钱逸斐1, 李倩玉1, 李松玲2, 覃雯莉1, 刘艳丰1( )
)
收稿日期:2024-04-05
接受日期:2025-06-06
出版日期:2025-09-28
发布日期:2025-09-30
通讯作者:
刘艳丰,研究员,博士;电子信箱:lyf7858188@163.com。基金资助:
ZHU Zijun1, QIAN Yife1, LI Qianyu1, LI Songling2, QIN Wenli1, LIU Yanfeng1( )
)
Received:2024-04-05
Accepted:2025-06-06
Online:2025-09-28
Published:2025-09-30
Contact:
LIU Yanfeng, E-mail: lyf7858188@163.com.Supported by:摘要:
目的·探究后期促进复合体亚基10(anaphase-promoting complex subunit 10,ANAPC10)在肝细胞癌(liver hepatocellular carcinoma,LIHC,常简称HCC)发生发展中的生物学功能及其潜在机制。方法·通过整合癌症基因组图谱数据库(The Cancer Genome Atlas,TCGA)中LIHC(TCGA_LIHC)、中国肝癌基因组计划数据库(China Hepatocellular Carcinoma Genome Project,CHCC)乙型肝炎病毒相关亚组(hepatitis B virus related subgroup,HBV)(CHCC_HBV)及基因表达综合数据库(Gene Expression Omnibus,GEO)的数据,分析ANAPC10在HCC中的表达模式,并在HCC细胞系中采用蛋白质印迹(Western blotting)和实时荧光定量PCR(quantitative real-time PCR,q-PCR)方法验证。通过shRNA介导的基因敲低实验,在MHCC-97H和SNU-398细胞系中下调ANAPC10的表达,探究ANAPC10的敲减与HCC细胞的体外增殖能力的关系。利用小鼠高压尾静脉注射技术构建Anapc10敲除原位肝癌模型,进一步证实肝脏ANAPC10的缺失对HCC发生发展的影响。对TCGA_LIHC和CHCC_HBV的RNA测序数据进行京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)、基因本体论(Gene Ontology,GO)通路富集分析。结果·ANAPC10在肿瘤组织中高表达,且其表达量与患者生存预后密切相关。体内外实验中下调ANAPC10可以有效抑制HCC的进展。ANAPC10主要通过影响PI3K-AKT-mTOR通路来对肿瘤的代谢进行重编程。在Anapc10敲除组的原位肝癌小鼠模型的肿瘤组织中,Akt和S6k的磷酸化水平降低;并且验证了下游脂代谢关键蛋白Fasn、Scd1的变化。结论·ANAPC10在HCC中高表达,与HCC的不良预后呈正相关,且促进HCC的发生及进展。ANAPC10激活PI3K-AKT-mTOR信号通路,促进HCC细胞的脂代谢重编程,最终促进肿瘤细胞的增殖与HCC的发生发展。
中图分类号:
朱子俊, 钱逸斐, 李倩玉, 李松玲, 覃雯莉, 刘艳丰. 后期促进复合体亚基10调控PI3K-AKT-mTOR通路促进肝细胞癌进展的研究[J]. 上海交通大学学报(医学版), 2025, 45(9): 1171-1182.
ZHU Zijun, QIAN Yife, LI Qianyu, LI Songling, QIN Wenli, LIU Yanfeng. Anaphase-promoting complex subunit 10 promotes hepatocellular carcinoma progression through regulation of the PI3K-AKT-mTOR signaling pathway[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(9): 1171-1182.
| shRNA | Target sequence (5'→3') | Loop sequence |
|---|---|---|
| shANAPC10-1 | CAGTCAGAGTAGGAAATAATT | CTCGAG |
| shANAPC10-2 | GACAATCATAAGAAGCCAACT | CTCGAG |
表1 shRNA序列(5'→3')
Tab 1 shRNA sequences (5'→3')
| shRNA | Target sequence (5'→3') | Loop sequence |
|---|---|---|
| shANAPC10-1 | CAGTCAGAGTAGGAAATAATT | CTCGAG |
| shANAPC10-2 | GACAATCATAAGAAGCCAACT | CTCGAG |
| sgRNA | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| sgAnacp10-1 | CACCAATCCGGCAACTTGAATTGG | AAACCCAATTCAAGTTGCCGGATT |
| sgAnacp10-2 | CACCGCAACTTGAATTGGTGGAAC | AAACGTTCCACCAATTCAAGTTGC |
| sgAnacp10-3 | CACCAGAAATCCGGCAACTTGA | AAACTCAAGTTGCCGGATTTCT |
表2 sgRNA序列(5'→3')
Tab 2 sgRNA sequences (5'→3')
| sgRNA | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| sgAnacp10-1 | CACCAATCCGGCAACTTGAATTGG | AAACCCAATTCAAGTTGCCGGATT |
| sgAnacp10-2 | CACCGCAACTTGAATTGGTGGAAC | AAACGTTCCACCAATTCAAGTTGC |
| sgAnacp10-3 | CACCAGAAATCCGGCAACTTGA | AAACTCAAGTTGCCGGATTTCT |
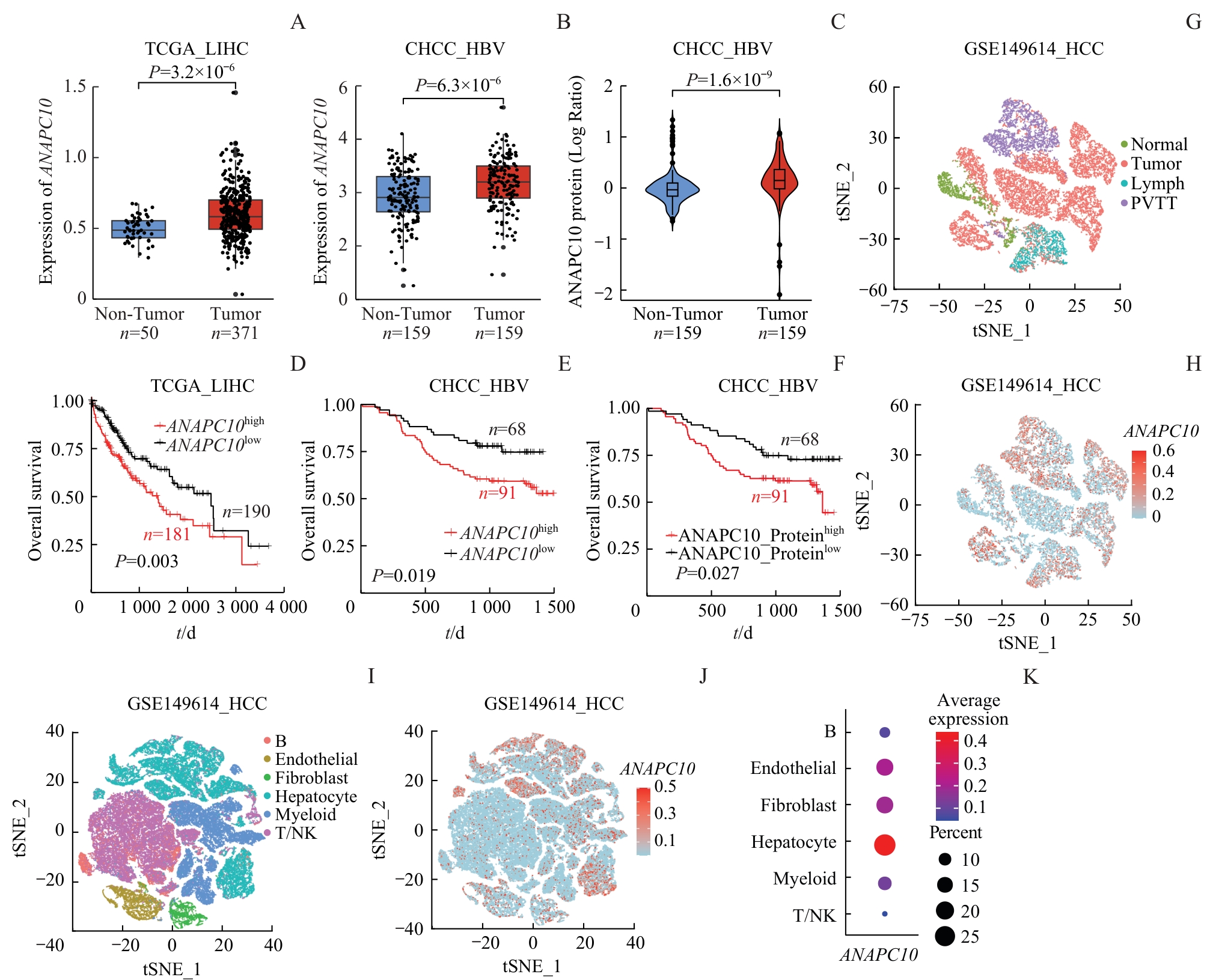
图1 ANAPC10 在HCC中的表达及与生存预后的关系Note: A/B. Comparison of ANAPC10 mRNA levels between HCC tumor tissues (tumor) and adjacent non-tumor tissues (non-tumor) in the TCGA_LIHC (A) and CHCC_HBV (B) datasets, analyzed by Student's t test. C. Comparison of ANAPC10 protein expression levels between HCC tumor tissues (Tumor) and adjacent non-tumor tissues (Non-Tumor) in the CHCC_HBV dataset, analyzed using Student's t-test. D/E. Kaplan-Meier survival analysis of overall survival (OS) stratified by high versus low ANAPC10 mRNA expression levels in the TCGA_LIHC (D) and CHCC_HBV (E) cohorts. F. Kaplan-Meier survival analysis of OS based on high versus low ANAPC10 protein expression levels in the CHCC_HBV cohort. G. t-SNE clustering plot of hepatocytes from different tissue sources (Normal, Tumor, Lymph, and PVTT) in the GEO single-cell RNA-seq dataset GSE149614. H. t-SNE clustering map displaying ANAPC10 expression levels in hepatocytes from HCC samples in the GSE149614 single-cell dataset. I. t-SNE clustering of different cell types (B cells, Endothelial cells, Fibroblasts, Hepatocytes, Myeloid cells, and T/NK cells) in HCC samples from the GSE149614 single-cell dataset. J. t-SNE clustering visualization of ANAPC10 expression across all cell populations in HCC samples from the GSE149614 dataset. K.Bubble chart illustrating ANAPC10 expression levels categorized by cell type (B cells, Endothelial cells, Fibroblasts, Hepatocytes, Myeloid cells, and T/NK cells) in the GSE149614 dataset.
Fig 1 Expression of ANAPC10 in hepatocellular carcinoma and its association with patient survival
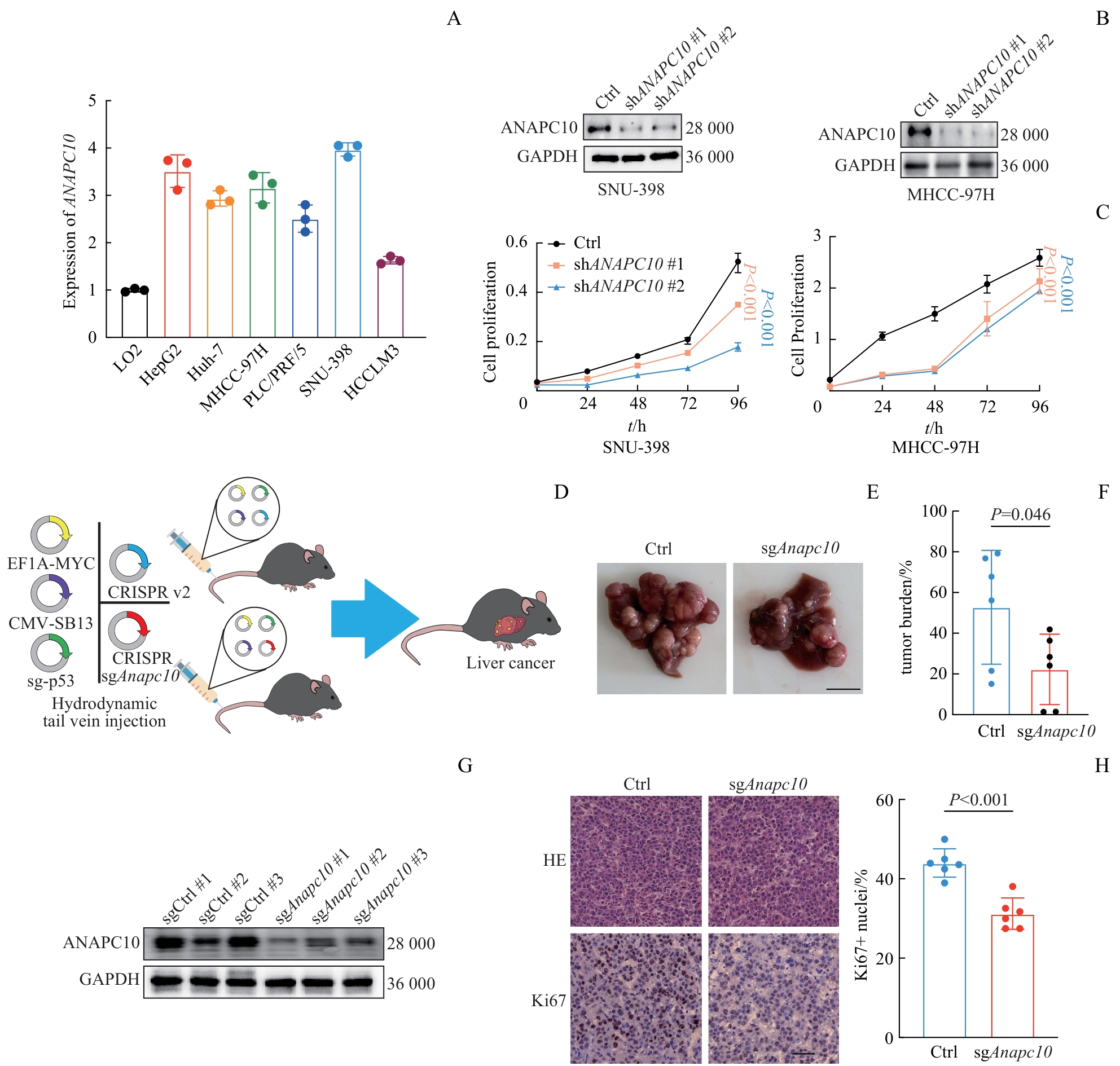
图2 ANAPC10 敲减肿瘤细胞的增殖情况及 Anapc10 敲除小鼠HCC的形成情况Note: A. qPCR was used to analyze the mRNA expression levels of ANAPC10 in various hepatocellular carcinoma cell lines (HepG2, Huh7, MHCC-97H, PLC/PRF/5, SNU-398, HCCLM3) relative to the normal hepatocyte cell line LO2. B. WB was employed to validate the protein levels of ANAPC10 after stable knockdown using shRNA in SNU-398 and MHCC-97H cell lines. C. A CCK-8 proliferation assay [with (n=6) replicates] was carried out in SNU-398 and MHCC-97H cell lines after stable ANAPC10 knockdown. Absorbance was measured at 450 nm (λ=450). D. An immunocompetent C57BL/6 mouse orthotopic HCC model was established by hydrodynamic tail-vein injection of plasmid solutions (2 mL in saline). The control group received a combination of 12 μg pT3-c-myc, 10 μg pX330-p53, 6.5 μg CMV-SB13, and 12 μg lentiCRISPR-v2. The knockout group received 12 μg pT3-c-myc, 10 μg pX330-p53, 6.5 μg CMV-SB13, and 12 μg sgAnapc10. E. Representative macroscopic images of livers with orthotopic HCC tumors were taken 34 d post-injection (n=6 per group, Ctrl vs sgAnapc10). Scale bar=10 mm. F. Hepatic tumor burden was quantified (n=6 per group, sgCtrl vs sgAnapc10). Tumor burden was calculated as the mean of (tumor area / liver area×100%). Statistical analysis was performed using Student's t-test. G. WB was used to validate the knockout efficiency of Anapc10 in tumor tissues (n=3 per group, sgCtrl vs sgAnapc10). Proteins were extracted from samples pulverized in liquid nitrogen. H. Representative H-E and Ki67 immunohistochemical staining of tumor tissues were conducted (scale bar=50 μm). Quantitative analysis of Ki67-positive cells was carried out (n=6 per group, Ctrl vs sgAnapc10). Statistical analysis was performed using Student's t-test.
Fig 2 Proliferation of tumor cells with ANAPC10 knockdown and HCC formation in Anapc10 knockout mice
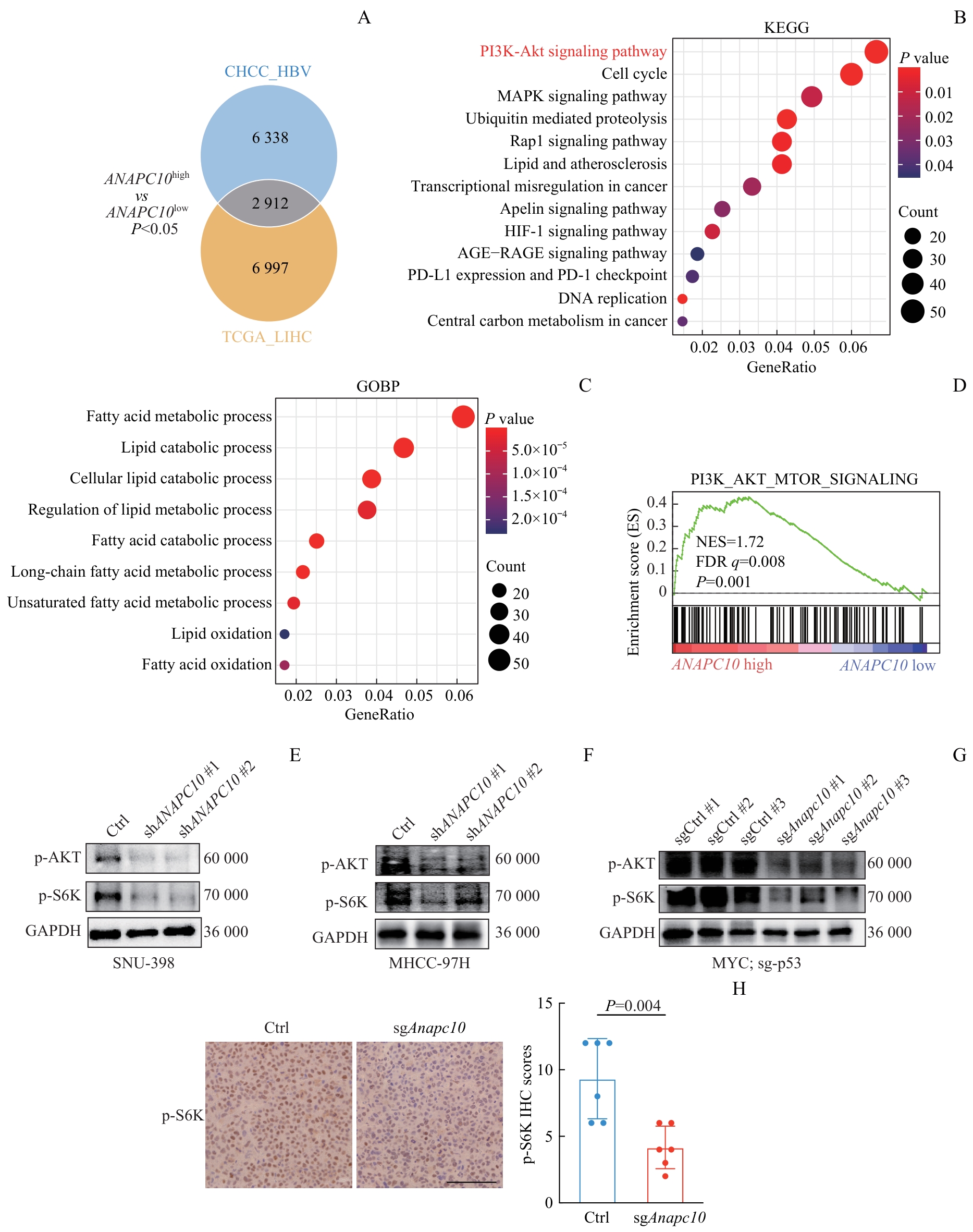
图3 ANAPC10 表达相关的通路富集分析及稳转细胞系和小鼠肝癌组织的WB和IHC验证Note:A. Patients from the TCGA_LIHC and CHCC_HBV cohorts were divided into ANAPC10-high-expression and ANAPC10-low-expression groups according to their mRNA levels. In the TCGA_LIHC cohort, there were 181 patients in the ANAPC10-high group and 190 in the ANAPC10-low group. In the CHCC_HBV cohort, there were 91 patients in the ANAPC10-high group and 68 in the ANAPC10-low group. Differentially expressed genes (DEGs) between the high- and low-expression groups (ranked by P value) were identified in both datasets, and 2 912 overlapping DEGs were found across the cohorts. B/C. KEGG (B) pathway and Gene Ontology Biological Process (GOBP) (C) enrichment analyses were carried out on the 2 912 common DEGs. D. Gene Set Enrichment Analysis (GSEA, version 4.3.2) was performed on CHCC_HBV patients stratified by ANAPC10 expression (91 patients in the ANAPC10-high group and 68 in the ANAPC10-low group). E/F. Protein expression levels of p-AKT and p-S6K in ANAPC10-knockdown SNU-398 (E) and MHCC-97H (F) cell lines were determined. G. Protein expression levels of p-Akt and p-S6k in the tumor tissues of the Anapc10-knockout orthotopic mouse hepatocellular carcinoma model were measured. H. Immunohistochemical (IHC) staining of p-S6k protein was conducted in the tumor tissues of the orthotopic hepatocellular carcinoma mouse model, along with relative IHC scoring statistics (n=6). Scale bar=100 µm. Statistical analysis was performed using the t-test.
Fig 3 Pathway enrichment analysis associated with ANAPC10 expression and WB/IHC validation in stable cell lines and mouse liver cancer tissues
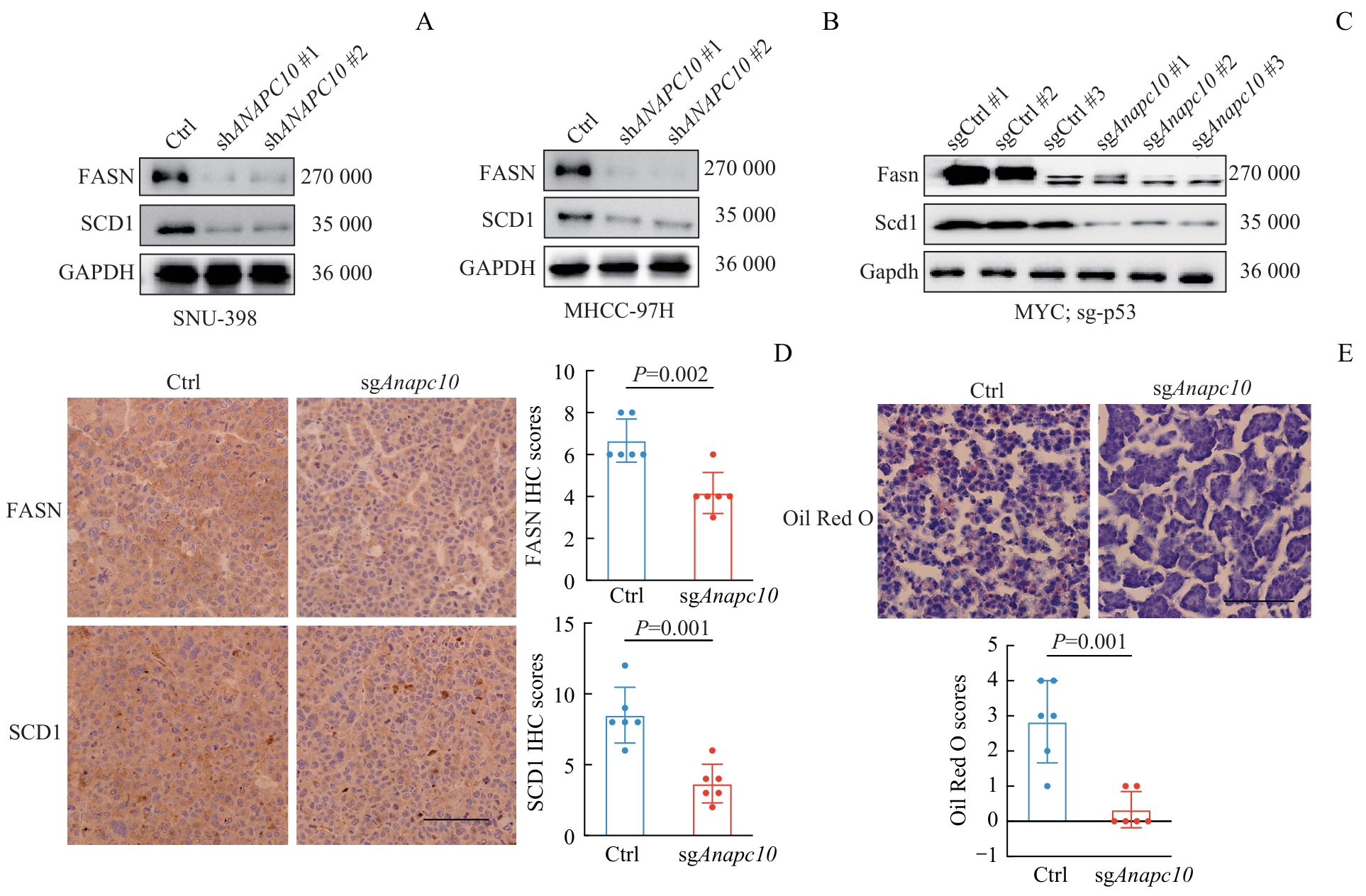
图4 稳转细胞系和小鼠肝癌组织中脂代谢关键蛋白的WB和IHC验证Note: A/B. Validation of changes in the protein expression of FASN and SCD1 in ANAPC10-knockdown stable cell lines (SNU-398 and MHCC-97H). C. Verification of the protein levels of Fasn and Scd1 in Anapc10-knockout orthotopic mouse HCC tumors. D. IHC staining of Fasn and Scd1 proteins in orthotopic HCC tumor tissues from mouse models, along with the corresponding quantitative IHC scores (n=6 biological replicates per group). E. Oil Red O staining of lipid droplets in the orthotopic HCC tumor tissues, accompanied by quantitative analysis of lipid accumulation (n=6 biological replicates per group). Scale bar=100 μm. Statistical analysis was carried out using the t-test.
Fig 4 WB and IHC validation of key lipid metabolism proteins in stable cell lines and murine hepatocellular carcinoma tissues
| [1] | BRAY F, LAVERSANNE M, SUNG H, et al. Global cancer statistics 2022: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2024, 74(3): 229-263. |
| [2] | MCGLYNN K A, PETRICK J L, THOMAS LONDON W. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability[J]. Clin Liver Dis, 2015, 19(2): 223-238. |
| [3] | ALLEMANI C, MATSUDA T, DI CARLO V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries[J]. Lancet, 2018, 391(10125): 1023-1075. |
| [4] | LLOVET J M, KELLEY R K, VILLANUEVA A, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2021, 7(1): 6. |
| [5] | BERTUCCIO P, TURATI F, CARIOLI G, et al. Global trends and predictions in hepatocellular carcinoma mortality[J]. J Hepatol, 2017, 67(2): 302-309. |
| [6] | SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249. |
| [7] | SINGAL A G, LAMPERTICO P, NAHON P. Epidemiology and surveillance for hepatocellular carcinoma: new trends[J]. J Hepatol, 2020, 72(2): 250-261. |
| [8] | CHEN J G, EGNER P A, NG D, et al. Reduced aflatoxin exposure presages decline in liver cancer mortality in an endemic region of China[J]. Cancer Prev Res (Phila), 2013, 6(10): 1038-1045. |
| [9] | HUANG D Q, EL-SERAG H B, LOOMBA R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention[J]. Nat Rev Gastroenterol Hepatol, 2021, 18(4): 223-238. |
| [10] | KOH J H, WANG M, SUZUKI H, et al. NAFLD and NAFLD-related HCC in Asia: burden and surveillance[J]. J Clin Exp Hepatol, 2024, 14(1): 101213. |
| [11] | HUANG D Q, SINGAL A G, KONO Y, et al. Changing global epidemiology of liver cancer from 2010 to 2019: Nash is the fastest growing cause of liver cancer[J]. Cell Metab, 2022, 34(7): 969-977.e2. |
| [12] | ZHOU J, SUN H C, WANG Z, et al. Guidelines for the diagnosis and treatment of hepatocellular carcinoma (2019 edition)[J]. Liver Cancer, 2020, 9(6): 682-720. |
| [13] | YAN K, DONG W, LI X W, et al. Early explosive recurrence of hepatocellular carcinoma after radical resection: risk factors and clinical significance[J]. Cancer Screen Prev, 2023, 2(4): 238-249. |
| [14] | POON R T, FAN S T. Evaluation of the new AJCC/UICC staging system for hepatocellular carcinoma after hepatic resection in Chinese patients[J]. Surg Oncol Clin N Am, 2003, 12(1): 35-50, viii. |
| [15] | LLOVET J M, RICCI S, MAZZAFERRO V, et al. Sorafenib in advanced hepatocellular carcinoma[J]. N Engl J Med, 2008, 359(4): 378-390. |
| [16] | KUDO M, FINN R S, QIN S K, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial[J]. Lancet, 2018, 391(10126): 1163-1173. |
| [17] | BRUIX J, QIN S K, MERLE P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial[J]. Lancet, 2017, 389(10064): 56-66. |
| [18] | ZHU A X, FINN R S, EDELINE J, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial[J]. Lancet Oncol, 2018, 19(7): 940-952. |
| [19] | FINN R S, QIN S K, IKEDA M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma[J]. N Engl J Med, 2020, 382(20): 1894-1905. |
| [20] | BARFORD D. Structure, function and mechanism of the anaphase promoting complex (APC/C)[J]. Q Rev Biophys, 2011, 44(2): 153-190. |
| [21] | HUANG S Y, WAN P, HUANG S Y, et al. The APC10 subunit of the anaphase-promoting complex/cyclosome orchestrates NLRP3 inflammasome activation during the cell cycle[J]. FEBS Lett, 2021, 595(19): 2463-2478. |
| [22] | WANG Y N, HAN T Y, GAN M X, et al. A novel function of anaphase promoting complex subunit 10 in tumor progression in non-small cell lung cancer[J]. Cell Cycle, 2019, 18(9): 1019-1032. |
| [23] | FAN H M, QUAN S X, YE Q, et al. A molecular framework underlying low-nitrogen-induced early leaf senescence in Arabidopsis thaliana[J]. Mol Plant, 2023, 16(4): 756-774. |
| [24] | GROSSBERGER R, GIEFFERS C, ZACHARIAE W, et al. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex[J]. J Biol Chem, 1999, 274(20): 14500-14507. |
| [25] | CARROLL C W, MORGAN D O. The Doc1 subunit is a processivity factor for the anaphase-promoting complex[J]. Nat Cell Biol, 2002, 4(11): 880-887. |
| [1] | 李倩玉, 钱逸斐, 李松玲, 朱子俊, 覃雯莉, 刘艳丰, 邱必军. Zeste 12抑制基因在肝细胞癌中的功能及机制[J]. 上海交通大学学报(医学版), 2025, 45(9): 1138-1148. |
| [2] | 许沐馨, 刘贤, 蒋立姗, 孙青. Nd:YAP激光生物刺激通过WNT/β-catenin信号通路促进人牙周韧带细胞增殖和成骨分化[J]. 上海交通大学学报(医学版), 2025, 45(5): 562-569. |
| [3] | 陈怡楠, 郑旸, 曾汉林, 雷鸣. Fas相关死亡结构域蛋白促进头颈部鳞状细胞癌细胞增殖能力的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(4): 404-414. |
| [4] | 张先洲, 杜凤麟, 吴雷, 任逸喆, 赵明娜, 娄加陶. OGT通过ERK信号通路促进非小细胞肺癌增殖的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(10): 1288-1297. |
| [5] | 李想, 魏鸣, 吴文曦, 罗小琴, 姚彪, 伍思宇. 芦丁对骨肉瘤生长和转移的体内外抑制作用[J]. 上海交通大学学报(医学版), 2025, 45(1): 20-28. |
| [6] | 孙晨玮, 海汪溪, 屈骞, 席云. [18F]F-FMISO和[18F]F-FLT PET/CT双核素显像预测胰腺癌耐药性的体内研究[J]. 上海交通大学学报(医学版), 2025, 45(1): 60-68. |
| [7] | 施灵玲, 程燕咏, 张磊. 七氟烷对原代少突胶质细胞增殖和分化的影响[J]. 上海交通大学学报(医学版), 2024, 44(9): 1115-1123. |
| [8] | 钱立恒, 温凯玲, 廖颖娜, 李书鑫, 聂惠贞. 分选链接蛋白1抑制结直肠癌细胞增殖和迁移的作用和机制研究[J]. 上海交通大学学报(医学版), 2024, 44(9): 1124-1135. |
| [9] | 谭露, 沈少明, 何平. 低氧诱导的长链非编码RNA 68在肝癌中的功能及其作用机制[J]. 上海交通大学学报(医学版), 2024, 44(6): 702-712. |
| [10] | 蔡人杰, 徐明. KHSRP通过ANK3调节前列腺癌细胞对雄激素的反应性[J]. 上海交通大学学报(医学版), 2024, 44(4): 417-426. |
| [11] | 安俊伊, 陈必颖, 陈循睿, 尹姗姗, 边洲亮, 刘峰. SFXN3在头颈部鳞状细胞癌中的表达及其对细胞增殖的影响[J]. 上海交通大学学报(医学版), 2024, 44(4): 427-434. |
| [12] | 李虎虓, 李笑甜, 赵旭日, 张桓瑜, 周薇, 宋忠臣. 牙龈素提取物对口腔鳞癌细胞HN6生物学特性的影响[J]. 上海交通大学学报(医学版), 2024, 44(2): 161-168. |
| [13] | 罗蓝鸽, 郑超, 雷鸣. 癌-睾丸抗原CT57促进肝癌细胞增殖、侵袭、迁移和上皮间质转化[J]. 上海交通大学学报(医学版), 2024, 44(11): 1335-1346. |
| [14] | 孔汝心, 周亚群, 魏婷宜, 雷鸣. 癌-睾丸抗原CT63在慢性髓系白血病中的作用及其机制[J]. 上海交通大学学报(医学版), 2024, 44(11): 1347-1358. |
| [15] | 高珂星, 廖春华, 李昇泽, 马双羽, 黄雷. 黏蛋白1调控肿瘤细胞恶性特征的功能位点分析[J]. 上海交通大学学报(医学版), 2024, 44(11): 1370-1382. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||