
上海交通大学学报(医学版) ›› 2024, Vol. 44 ›› Issue (11): 1370-1382.doi: 10.3969/j.issn.1674-8115.2024.11.004
• 论著 · 基础研究 • 上一篇
收稿日期:2024-02-11
接受日期:2024-03-22
出版日期:2024-11-28
发布日期:2024-11-28
通讯作者:
黄雷
E-mail:xingke_gao@163.com;leihuang@shsmu.edu.cn
作者简介:高珂星(2000—),女,硕士生;电子信箱:xingke_gao@163.com。
基金资助:
GAO Kexing( ), LIAO Chunhua, LI Shengze, MA Shuangyu, HUANG Lei(
), LIAO Chunhua, LI Shengze, MA Shuangyu, HUANG Lei( )
)
Received:2024-02-11
Accepted:2024-03-22
Online:2024-11-28
Published:2024-11-28
Contact:
HUANG Lei
E-mail:xingke_gao@163.com;leihuang@shsmu.edu.cn
Supported by:摘要:
目的·探究黏蛋白1(mucin 1,MUC1)调控肿瘤细胞增殖、迁移和干性维持的功能位点。方法·通过癌症基因组图谱(The Cancer Genome Atlas,TCGA)数据库分析寻找MUC1基因在不同癌症中的突变特征,对不同MUC1突变位点进行分析及定位,并按突变出现频率排序;通过Western blotting筛选出突变频率较高且蛋白稳定表达的MUC1突变体,利用乳腺癌细胞株BT549敲除MUC1细胞系和乳腺非转化细胞株MCF-10A,应用慢病毒表达系统构建MUC1野生型(MUC1-WT)和突变体稳定表达的细胞系。采用免疫荧光法检测不同MUC1突变体的细胞定位。以MUC1-WT为阳性对照、MUC1-AQA功能丧失突变体为阴性对照,对不同突变细胞的肿瘤生物学功能进行分析:通过细胞计数试剂盒-8(cell counting kit-8,CCK-8)及克隆形成实验检测细胞增殖能力;通过划痕实验及Transwell实验检测细胞迁移能力;通过成球实验检测细胞干性。使用PyMOL软件分析MUC1突变体结构定位并通过蛋白质对接软件(ZDOCK Server)进行分子对接分析。结果·在TCGA数据库中得到102个位于MUC1编码区的突变,其中P418S、S251R、V359I、N271S、N465H 5个错义突变出现频率较高且位于非数目可变串联重复序列(non-variable number of tandem repeats,non-VNTR)区域。进一步检测发现MUC1-S251R、N271S、V359I突变体可稳定表达;细胞定位分析发现这3个突变体主要分布于细胞质,同时细胞核也有一定的分布,核质比与野生型未见明显差异。表达不同MUC1突变体细胞的肿瘤生物学功能分析发现:① MUC1-WT高表达显著增强BT549和MCF-10A细胞的增殖能力;与MUC1-WT细胞相比,MUC1-AQA、S251R、N271S突变体细胞增殖能力下降,但MUC1-V359I突变体细胞与MUC1-WT细胞具有相似的增殖能力。② MUC1-WT高表达细胞的迁移能力显著增强,而MUC1-AQA细胞迁移能力减弱。在BT549细胞中,MUC1-S251R与MUC1-V359I突变体细胞迁移能力与MUC1-WT细胞相似,但MUC1-N271S细胞的迁移能力较MUC1-WT细胞降低。在MCF-10A细胞中,MUC1-N271S与MUC1-V359I细胞的迁移能力接近MUC1-WT细胞;但MUC1-S251R细胞较MUC1-WT细胞迁移能力显著下降。③ MUC1-WT高表达显著增强2种细胞的干性,而MUC1-AQA细胞干性丧失;MUC1-N271S、V359I与MUC1-WT具有相似的维持细胞干性的能力,而MUC1-S251R使细胞干性减弱。PyMOL软件分析结果显示,MUC1-N271S及V359I位于海胆精子蛋白-肠激酶-聚集蛋白(sperm protein-enterokinase-agarin,SEA)区域及附近,分别处于loop区及β-折叠处;分子对接结果显示,MUC1-WT及V359I与表皮生长因子受体(epidermal growth factor receptor,EGFR)胞外域形成复合物的稳定性强于MUC1-N271S和S251R,其稳定性排序为V359I>WT>N271S>S251R。结论·MUC1突变体对肿瘤细胞的生物学功能具有不同影响,其对增殖能力影响可能与EGFR信号通路相关。MUC1-V359I与MUC1-WT相似,并未影响MUC1对肿瘤细胞增殖、迁移及干性维持的作用;而MUC1-S251、N271位点可能参与细胞增殖和迁移的信号通路调控且MUC1-S251位点对维持细胞干性较为重要。
中图分类号:
高珂星, 廖春华, 李昇泽, 马双羽, 黄雷. 黏蛋白1调控肿瘤细胞恶性特征的功能位点分析[J]. 上海交通大学学报(医学版), 2024, 44(11): 1370-1382.
GAO Kexing, LIAO Chunhua, LI Shengze, MA Shuangyu, HUANG Lei. Functional site analysis of mucin 1 in regulating the malignant characteristics of tumor cells[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(11): 1370-1382.
| Primer | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| AQA | GCCTTGGCTGTCGCTCAGGCCCGCCGAAAGAAC | GTTCTTTCGGCGGGCCTGAGCGACAGCCAAGGC |
| S251R | CCTCCAATCACAGGACTTCTCCCCAG | CTGGGGAGAAGTCCTGTGATTGGAGG |
| N271S | TTCACATTTCAAGCCTCCAGTTTAATT | AATTAAACTGGAGGCTTGAAATGTGAA |
| V359I | ACGATCTCAGACATCAGCGTGAGTGAT | ATCACTCACGCTTACGTCTGAGATCGT |
| P418S | CAGCTGGACATCTTTTCAGCCCGGGAT | ATCCCGGGCTGAAAAGATGTCCAGCTG |
| N465H | TCTTACACACACCCAGCAGTGGCAGCC | GGCTGCCACTGCTGGGTGTGTGTAAGA |
表1 PCR引物序列
Tab 1 Primer sequences for PCR
| Primer | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| AQA | GCCTTGGCTGTCGCTCAGGCCCGCCGAAAGAAC | GTTCTTTCGGCGGGCCTGAGCGACAGCCAAGGC |
| S251R | CCTCCAATCACAGGACTTCTCCCCAG | CTGGGGAGAAGTCCTGTGATTGGAGG |
| N271S | TTCACATTTCAAGCCTCCAGTTTAATT | AATTAAACTGGAGGCTTGAAATGTGAA |
| V359I | ACGATCTCAGACATCAGCGTGAGTGAT | ATCACTCACGCTTACGTCTGAGATCGT |
| P418S | CAGCTGGACATCTTTTCAGCCCGGGAT | ATCCCGGGCTGAAAAGATGTCCAGCTG |
| N465H | TCTTACACACACCCAGCAGTGGCAGCC | GGCTGCCACTGCTGGGTGTGTGTAAGA |
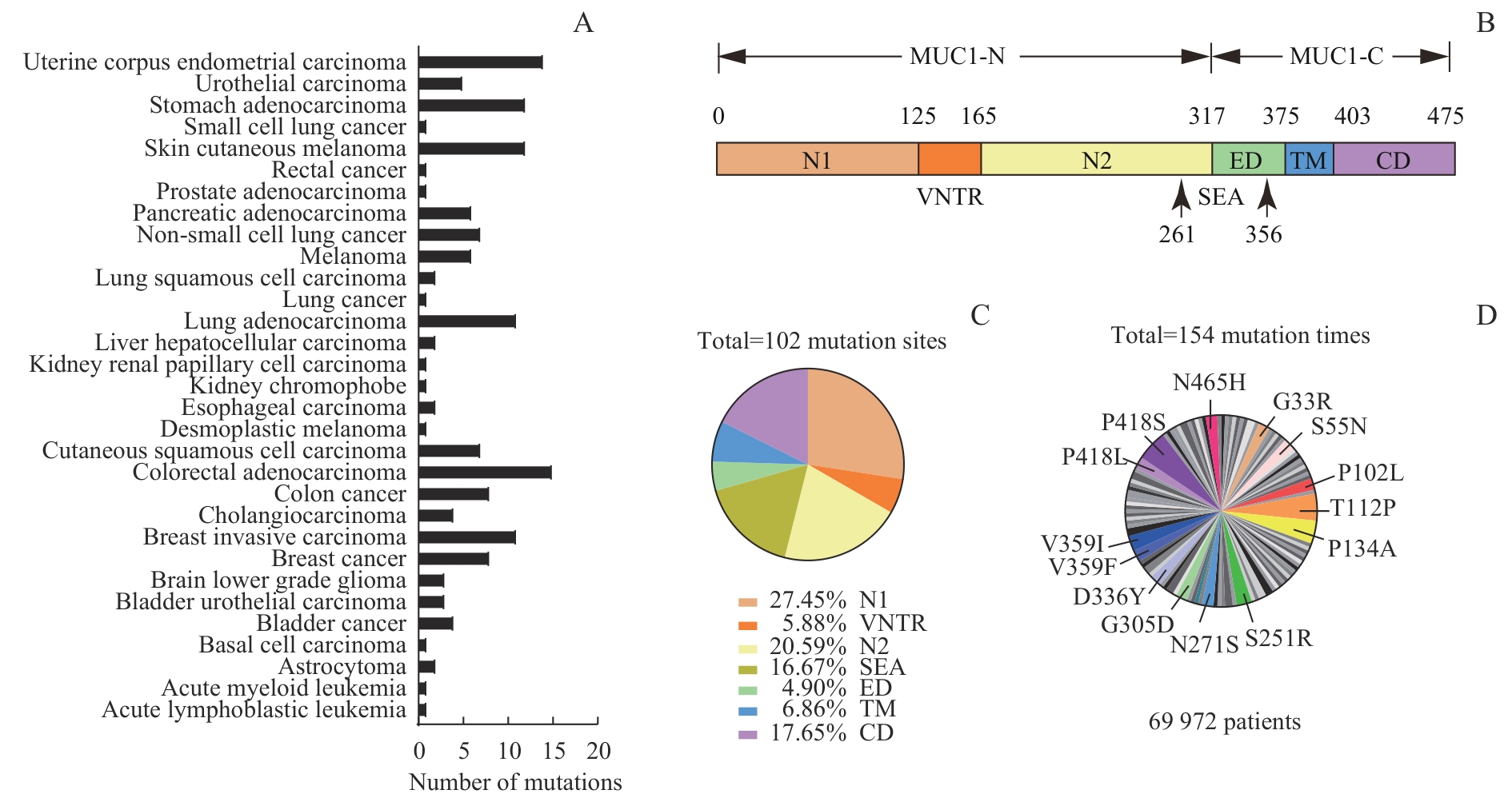
图1 TCGA数据库中 MUC1 突变位点来源、定位及频率分析Note: A. Proportion of MUC1 mutations in various tumor types. B. MUC1 protein consisting of N1, VNTR, N2, SEA, ED, TM, and CD domains. C. Location of 102 mutation sites in MUC1 protein. D. The mutation sites with a frequency of 1.95% or more in tumor samples.
Fig 1 Analysis of sources, positions, and frequencies of MUC1 mutation sites in the TCGA database
| Mutant | Cancer type | Location | Frequency/% |
|---|---|---|---|
| P418S | Lung squamous cell carcinoma | CD | 5.19 |
| T112P | Bladder urothelial carcinoma/pancreatic adenocarcinoma | N1 | 4.55 |
| P134A | Pancreatic adenocarcinoma/melanoma | VNTR | 3.90 |
| S251R | Esophagogastric cancer/stomach adenocarcinoma | N2 | 2.60 |
| V359I | Colorectal adenocarcinoma/breast invasive carcinoma | ED | 2.60 |
| G33R | Cutaneous squamous cell carcinoma | N1 | 1.95 |
| S55N | Uterine corpus endometrial carcinoma | N1 | 1.95 |
| P102L | Stomach adenocarcinoma | N1 | 1.95 |
| N271S | Breast invasive carcinoma | SEA | 1.95 |
| G305D | Stomach adenocarcinoma | SEA | 1.95 |
| D336Y | Lung adenocarcinoma | SEA | 1.95 |
| V359F | Breast invasive carcinoma | ED | 1.95 |
| P418L | Cutaneous squamous cell carcinoma | CD | 1.95 |
| N465H | Uterine corpus endometrioid carcinoma | CD | 1.95 |
表2 14个 MUC1 高频突变体的信息
Tab 2 Information of 14 MUC1 mutants with high frequency
| Mutant | Cancer type | Location | Frequency/% |
|---|---|---|---|
| P418S | Lung squamous cell carcinoma | CD | 5.19 |
| T112P | Bladder urothelial carcinoma/pancreatic adenocarcinoma | N1 | 4.55 |
| P134A | Pancreatic adenocarcinoma/melanoma | VNTR | 3.90 |
| S251R | Esophagogastric cancer/stomach adenocarcinoma | N2 | 2.60 |
| V359I | Colorectal adenocarcinoma/breast invasive carcinoma | ED | 2.60 |
| G33R | Cutaneous squamous cell carcinoma | N1 | 1.95 |
| S55N | Uterine corpus endometrial carcinoma | N1 | 1.95 |
| P102L | Stomach adenocarcinoma | N1 | 1.95 |
| N271S | Breast invasive carcinoma | SEA | 1.95 |
| G305D | Stomach adenocarcinoma | SEA | 1.95 |
| D336Y | Lung adenocarcinoma | SEA | 1.95 |
| V359F | Breast invasive carcinoma | ED | 1.95 |
| P418L | Cutaneous squamous cell carcinoma | CD | 1.95 |
| N465H | Uterine corpus endometrioid carcinoma | CD | 1.95 |
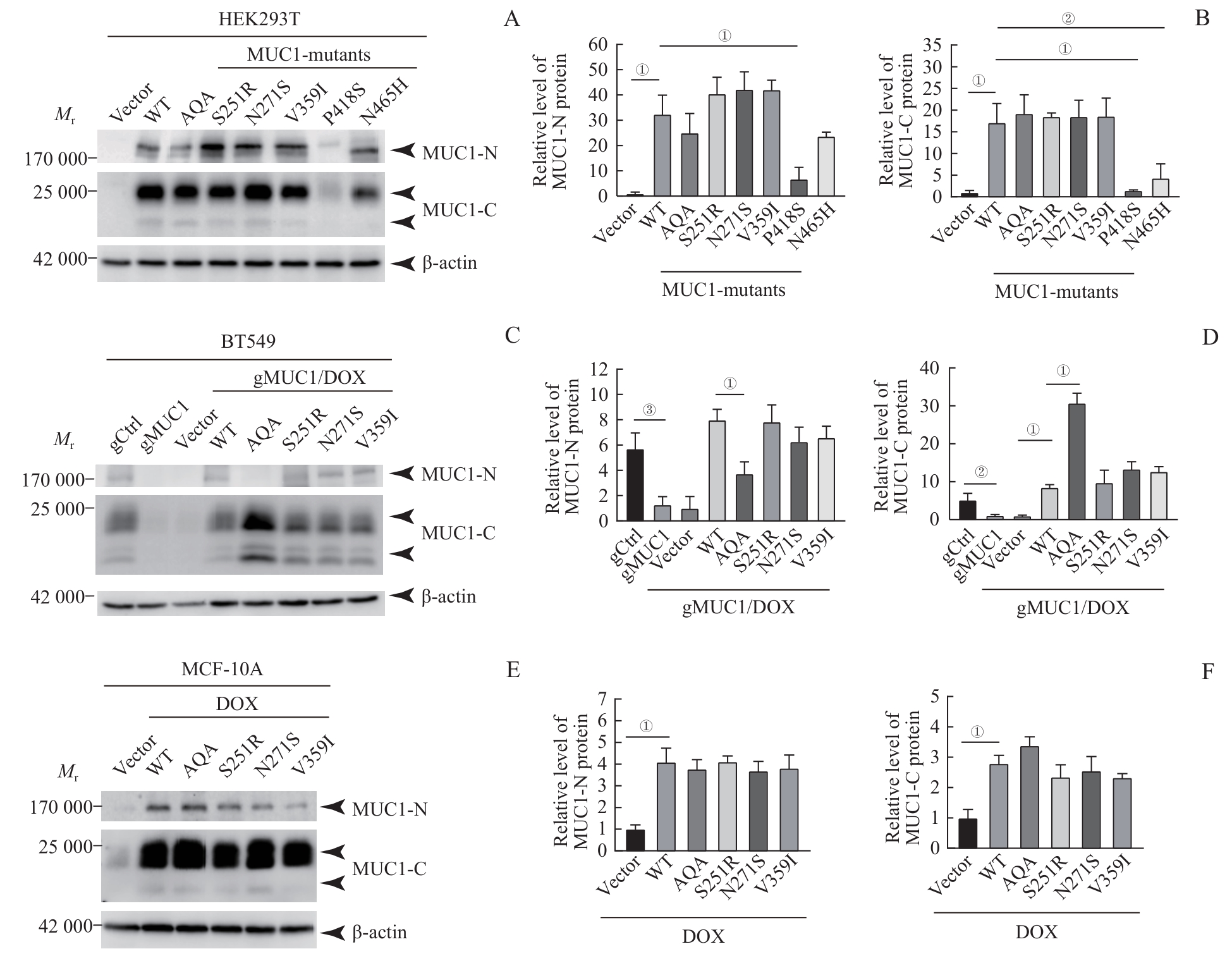
图2 MUC1-WT及其突变体在细胞中的蛋白表达水平Note: A/C/E. Western blotting was used to detect MUC1 protein expression levels in HEK293T cells (A), BT549 cells (C) and MCF-10A cells (E). B/D/F. Statistical analysis of MUC1 protein levels in HEK293T cells (B), BT549 cells (D) and MCF-10A cells (F). ①P<0.001, ②P=0.020, ③P=0.002.
Fig 2 Protein expression levels of MUC1-WT and mutants in the cells
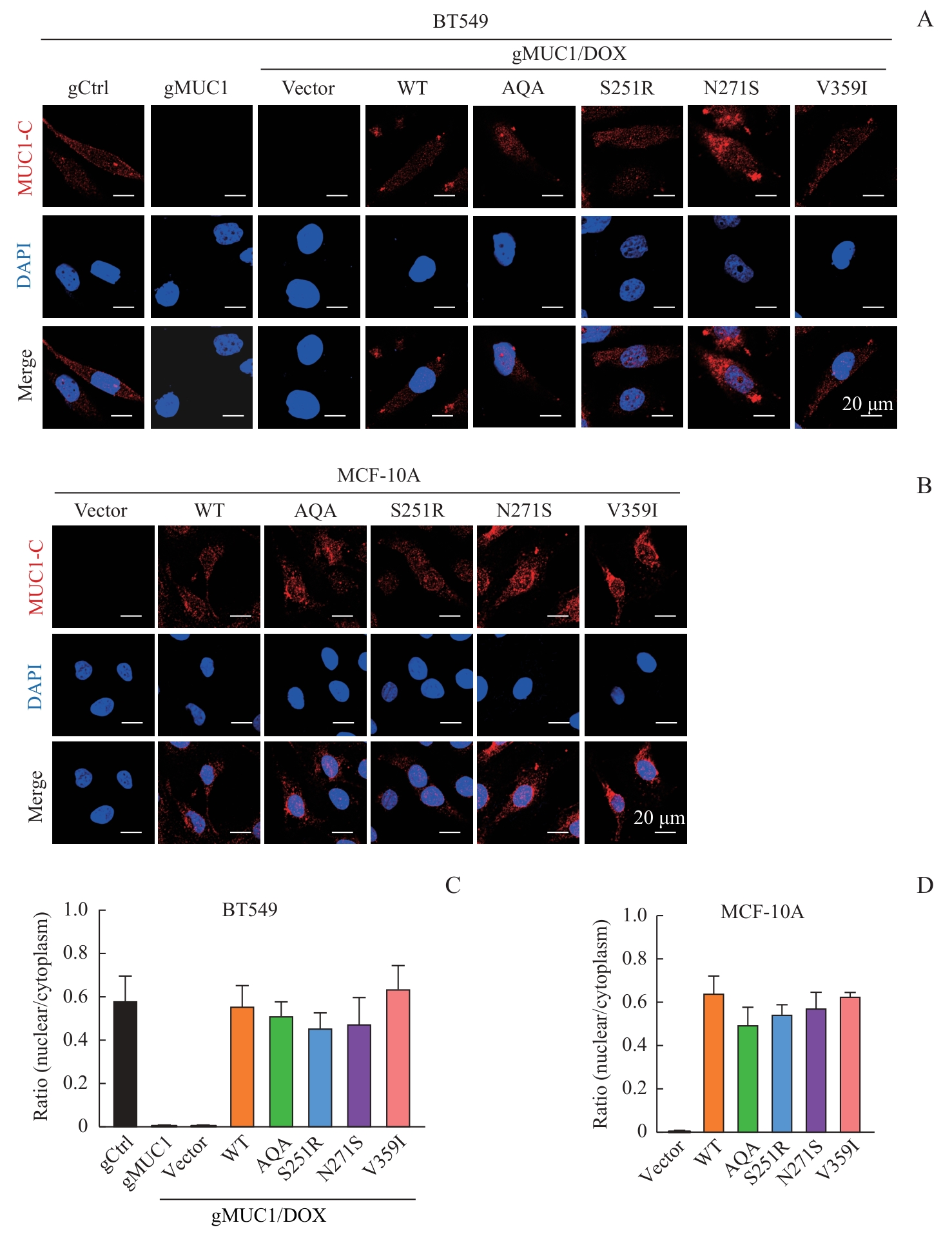
图3 MUC1-WT及其突变体的细胞定位Note: A/B. Detection of cellular localization of MUC1 mutants by immunofluorescence in BT549 cells (A) and MCF-10A cells (B)(×600). C/D. Statistical analysis of MUC1 nuclear/cytoplasm ratio in BT549 cells (C) and MCF-10A cells (D).
Fig 3 Cellular localization of MUC1-WT and mutants
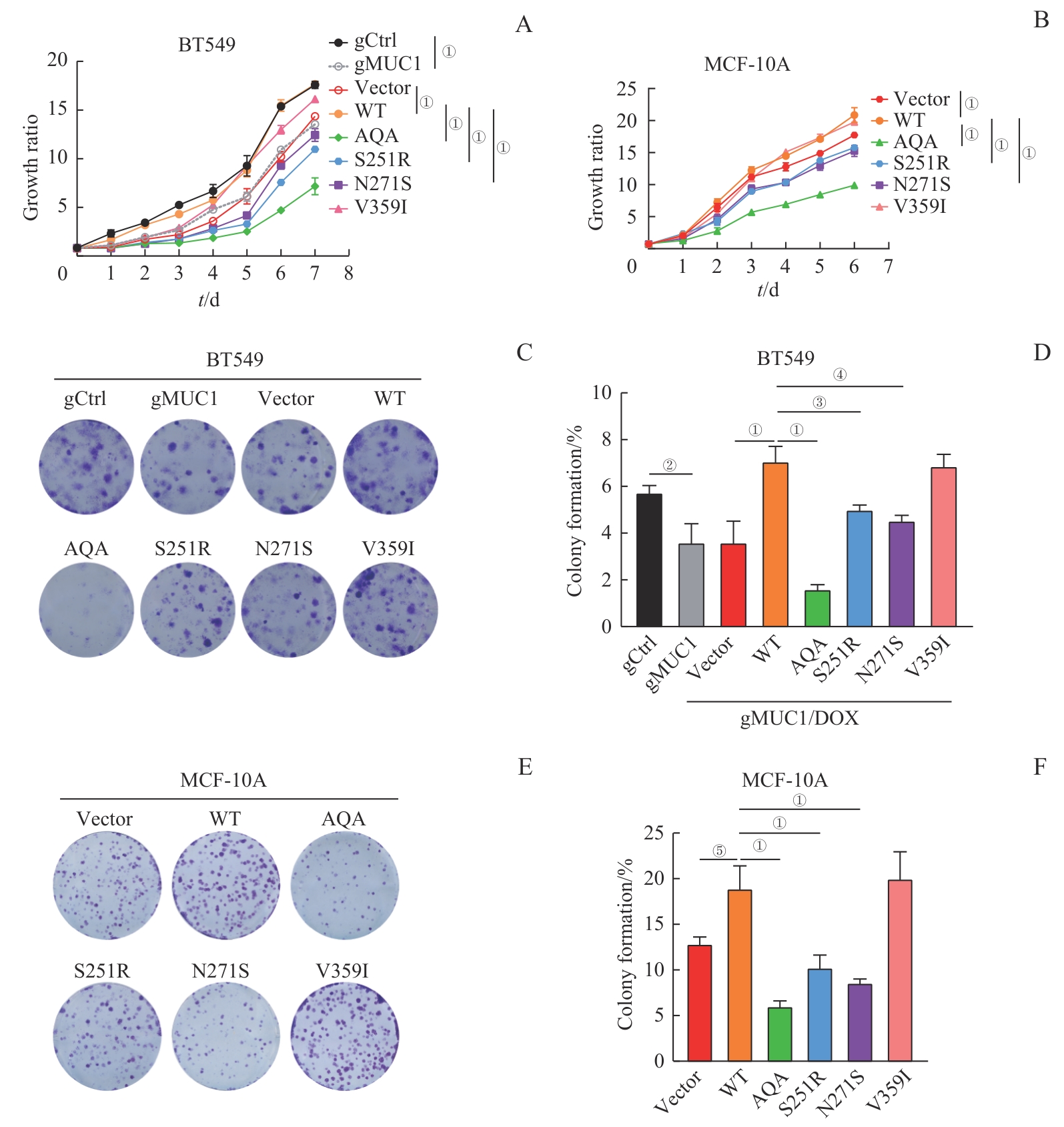
图4 MUC1-WT及其突变体对细胞增殖能力的影响Note: A/B. Growth curves of BT549 cells (A) and MCF-10A cells (B) expressing MUC1-WT or mutants detected by CCK-8. C/D. Colony formation analysis of BT549 cells expressing MUC1-WT or mutants. Representative crystal violet-stained images (C) and corresponding statistical analysis (D). E/F. Colony formation analysis of MCF-10A cells expressing MUC1-WT or mutants. Representative crystal violet-stained images (E) and corresponding statistical analysis (F). ①P<0.001, ②P=0.003, ③P=0.005, ④P=0.007, ⑤P=0.009.
Fig 4 Effect of MUC1-WT and mutants on cell proliferation
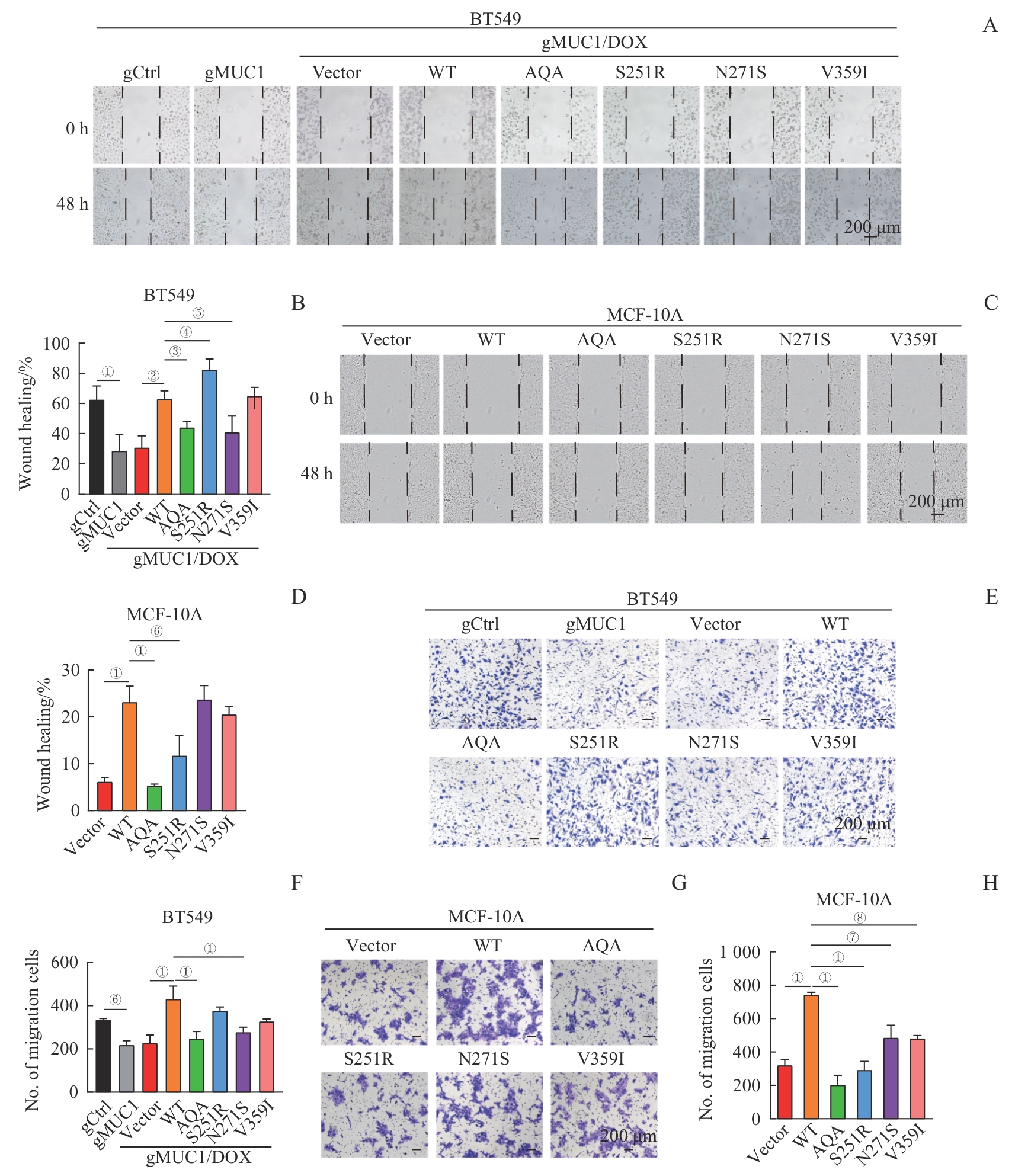
图5 MUC1-WT及其突变体对细胞迁移的影响Note: A/B. Wound healing assay of BT549 cells expressing MUC1-WT or mutants. Representative images of cells at 0 h and 48 h (A, ×200) and corresponding statistical analysis (B). C/D. Wound healing assay of MCF-10A cells expressing MUC1-WT or mutants. Representative images of cells at 0 h and 48 h (C, ×200) and corresponding statistical analysis (D). E/F. Transwell migration analysis of BT549 cells expressing MUC1-WT or mutants. Representative crystal violet-stained images (E, ×100) and corresponding statistical analysis (F). G/H. Transwell migration analysis of MCF-10A cells expressing MUC1-WT or mutants. Representative crystal violet-stained images (G, ×100) and corresponding statistical analysis (H). ①P<0.001, ②P=0.004, ③P=0.014, ④P=0.030, ⑤P=0.025, ⑥P=0.003, ⑦P=0.050, ⑧P=0.042.
Fig 5 Effect of MUC1-WT and mutants on cell migration
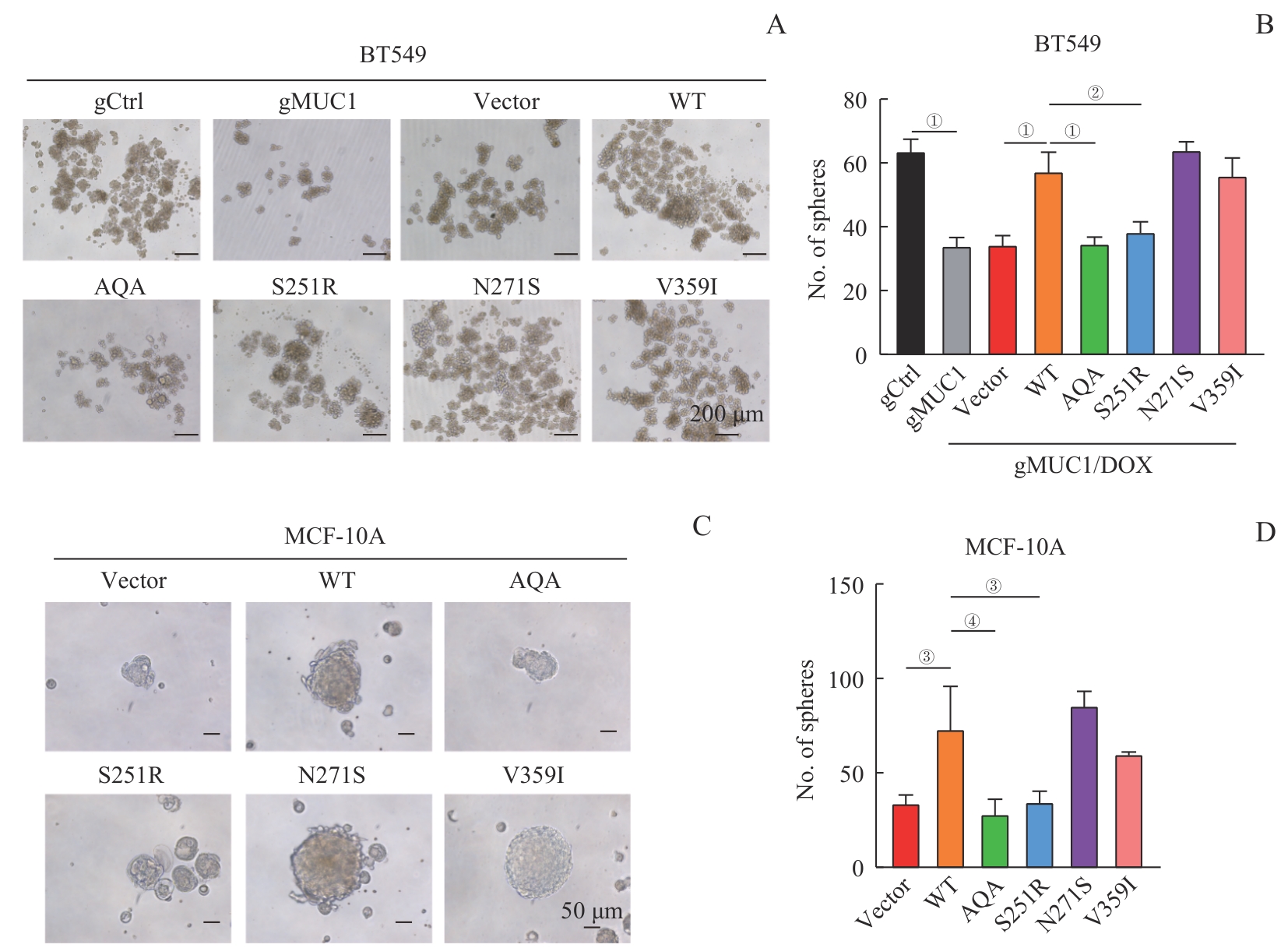
图6 MUC1-WT及其突变体对细胞干性的影响Note: A/B. Sphere formation assay of BT549 cells expressing MUC1-WT or mutants. Representative images of spheres (A, ×100) and corresponding statistical analysis (B). C/D. Sphere formation assay of MCF-10A cells expressing MUC1-WT or mutants. Representative images of spheres (C, ×200) and corresponding statistical analysis (D). ①P<0.001, ②P=0.004, ③P=0.007, ④P=0.002.
Fig 6 Effect of MUC1-WT and mutants on cell stemness
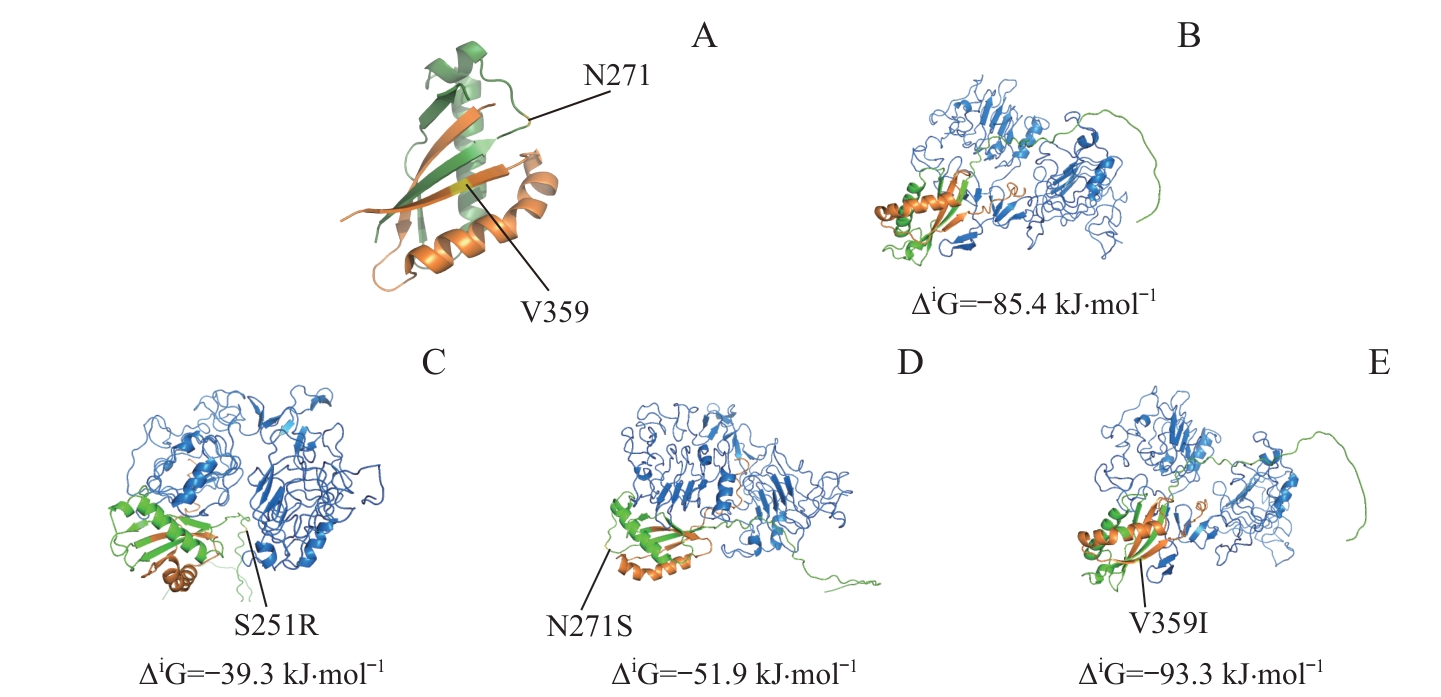
图7 SEA区域及附近的MUC1突变定位及相应突变体与EGFR-ECD的分子对接分析Note: A. PyMoL software was used to localize MUC1 mutants in or around the SEA domain. B?E. ZDOCK Server was used to analyze the molecular docking of MUC1-WT (B) and mutants (C, S251R; D, N271S; E, V359I) with EGFR-ECD. Green represents the MUC1-N region (before the GSVVV sequence), orange represents the MUC1-C region (after the GSVVV sequence), blue represents EGFR-ECD region, and yellow represents the localization of the mutation sites.
Fig 7 Analysis of localization of MUC1 mutation sites in or around the SEA domain and molecular docking of the mutants with EGFR-ECD
| 1 | KUFE D W. Mucins in cancer: function, prognosis and therapy[J]. Nat Rev Cancer, 2009, 9(12): 874-885. |
| 2 | ANDRIANIFAHANANA M, MONIAUX N, BATRA S K. Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases[J]. Biochim Biophys Acta, 2006, 1765(2): 189-222. |
| 3 | MONIAUX N, ANDRIANIFAHANANA M, BRAND R E, et al. Multiple roles of mucins in pancreatic cancer, a lethal and challenging malignancy[J]. Br J Cancer, 2004, 91(9): 1633-1638. |
| 4 | GAEMERS I C, VOS H L, VOLDERS H H, et al. A STAT-responsive element in the promoter of the episialin/MUC1 gene is involved in its overexpression in carcinoma cells[J]. J Biol Chem, 2001, 276(9): 6191-6199. |
| 5 | SHENG Y H, LOURIE R, LINDÉN S K, et al. The MUC13 cell-surface mucin protects against intestinal inflammation by inhibiting epithelial cell apoptosis[J]. Gut, 2011, 60(12): 1661-1670. |
| 6 | SINGH A P, MONIAUX N, CHAUHAN S C, et al. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis[J]. Cancer Res, 2004, 64(2): 622-630. |
| 7 | NATH S, MUKHERJEE P. MUC1: a multifaceted oncoprotein with a key role in cancer progression[J]. Trends Mol Med, 2014, 20(6): 332-342. |
| 8 | CHEN W Q, ZHANG Z, ZHANG S Q, et al. MUC1: structure, function, and clinic application in epithelial cancers[J]. Int J Mol Sci, 2021, 22(12): 6567. |
| 9 | HATTRUP C L, GENDLER S J. Structure and function of the cell surface (tethered) mucins[J]. Annu Rev Physiol, 2008, 70: 431-457. |
| 10 | LEVITIN F, STERN O, WEISS M, et al. The MUC1 SEA module is a self-cleaving domain[J]. J Biol Chem, 2005, 280(39): 33374-33386. |
| 11 | CARSON D D. The cytoplasmic tail of MUC1: a very busy place[J]. Sci Signal, 2008, 1(27): pe35. |
| 12 | KUFE D W. MUC1-C in chronic inflammation and carcinogenesis; emergence as a target for cancer treatment[J]. Carcinogenesis, 2020, 41(9): 1173-1183. |
| 13 | LENG Y M, CAO C, REN J, et al. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62[J]. J Biol Chem, 2007, 282(27): 19321-19330. |
| 14 | RAINA D, AHMAD R, RAJABI H, et al. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells[J]. Int J Oncol, 2012, 40(5): 1643-1649. |
| 15 | VAN PUTTEN J P M, STRIJBIS K. Transmembrane mucins: signaling receptors at the intersection of inflammation and cancer[J]. J Innate Immun, 2017, 9(3): 281-299. |
| 16 | YOLKEN R H, PETERSON J A, VONDERFECHT S L, et al. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis[J]. J Clin Invest, 1992, 90(5): 1984-1991. |
| 17 | LAU S K, WEISS L M, CHU P G. Differential expression of MUC1, MUC2, and MUC5AC in carcinomas of various sites: an immunohistochemical study[J]. Am J Clin Pathol, 2004, 122(1): 61-69. |
| 18 | KASHYAP B, KULLAA A M. Regulation of mucin 1 expression and its relationship with oral diseases[J]. Arch Oral Biol, 2020, 117: 104791. |
| 19 | SAFI F, KOHLER I, RÖTTINGER E, et al. The value of the tumor marker CA 15-3 in diagnosing and monitoring breast cancer[J]. Cancer, 1991, 68(3): 574-582. |
| 20 | STEINBERG W. The clinical utility of the CA 19-9 tumor-associated antigen[J]. Am J Gastroenterol, 1990, 85(4): 350-355. |
| 21 | SUPRUNIUK K, RADZIEJEWSKA I. MUC1 is an oncoprotein with a significant role in apoptosis (Review)[J]. Int J Oncol, 2021, 59(3): 68. |
| 22 | YAMAMOTO S, KAIMORI J Y, YOSHIMURA T, et al. Analysis of an ADTKD family with a novel frameshift mutation in MUC1 reveals characteristic features of mutant MUC1 protein[J]. Nephrol Dial Transplant, 2017, 32(12): 2010-2017. |
| 23 | WENZEL A, ALTMUELLER J, EKICI A B, et al. Single molecule real time sequencing in ADTKD-MUC1 allows complete assembly of the VNTR and exact positioning of causative mutations[J]. Sci Rep, 2018, 8(1): 4170. |
| 24 | LI Q F, CHU Y K, LI S Z, et al. The oncoprotein MUC1 facilitates breast cancer progression by promoting Pink1-dependent mitophagy via ATAD3A destabilization[J]. Cell Death Dis, 2022, 13(10): 899. |
| 25 | JIN W, LIAO X D, LV Y P, et al. MUC1 induces acquired chemoresistance by upregulating ABCB1 in EGFR-dependent manner[J]. Cell Death Dis, 2017, 8(8): e2980. |
| 26 | RAZAWI H, KINLOUGH C L, STAUBACH S, et al. Evidence for core 2 to core 1 O-glycan remodeling during the recycling of MUC1[J]. Glycobiology, 2013, 23(8): 935-945. |
| 27 | KINLOUGH C L, MCMAHAN R J, POLAND P A, et al. Recycling of MUC1 is dependent on its palmitoylation[J]. J Biol Chem, 2006, 281(17): 12112-12122. |
| 28 | PASTRELLO C, SANTAROSA M, FORNASARIG M, et al. MUC gene abnormalities in sporadic and hereditary mucinous colon cancers with microsatellite instability[J]. Dis Markers, 2005, 21(3): 121-126. |
| 29 | ZHANG L X, VLAD A, MILCAREK C, et al. Human mucin MUC1 RNA undergoes different types of alternative splicing resulting in multiple isoforms[J]. Cancer Immunol Immunother, 2013, 62(3): 423-435. |
| 30 | STRATTON M R, CAMPBELL P J, FUTREAL P A. The cancer genome[J]. Nature, 2009, 458(7239): 719-724. |
| 31 | MARTÍNEZ-JIMÉNEZ F, MUIÑOS F, SENTÍS I, et al. A compendium of mutational cancer driver genes[J]. Nat Rev Cancer, 2020, 20(10): 555-572. |
| 32 | NABAVINIA M S, GHOLOOBI A, CHARBGOO F, et al. Anti-MUC1 aptamer: a potential opportunity for cancer treatment[J]. Med Res Rev, 2017, 37(6): 1518-1539. |
| 33 | HOU Y, GAO J, XU H, et al. PPARγ E3 ubiquitin ligase regulates MUC1-C oncoprotein stability[J]. Oncogene, 2014, 33(49): 5619-5625. |
| 34 | LIAO C H, YU L P, PANG Z, et al. WWP1 targeting MUC1 for ubiquitin-mediated lysosomal degradation to suppress carcinogenesis[J]. Signal Transduct Target Ther, 2021, 6(1): 297. |
| 35 | ZHANG W B, LIU M W, YU L L, et al. Perturbation effect of single polar group substitution on the self-association of amphiphilic peptide helices[J]. J Colloid Interface Sci, 2022, 610: 1005-1014. |
| 36 | ZHANG W B, LIU M W, WANG Y, et al. β-sheet assembly translates conservative single-site mutation into a perturbation in macroscopic structure[J]. Nano Lett, 2023, 23(6): 2370-2378. |
| 37 | MACAO B, JOHANSSON D G, HANSSON G C, et al. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin[J]. Nat Struct Mol Biol, 2006, 13(1): 71-76. |
| 38 | MERLIN J, STECHLY L, DE BEAUCÉ S, et al. Galectin-3 regulates MUC1 and EGFR cellular distribution and EGFR downstream pathways in pancreatic cancer cells[J]. Oncogene, 2011, 30(22): 2514-2525. |
| [1] | 木司塔巴·木台力甫, 王俊杰, 钱云臻, 陈溯源, 邵达, 张志刚, 李冬雪. 预免疫策略结合mVenus-p27K-系统构建休眠肿瘤小鼠模型[J]. 上海交通大学学报(医学版), 2024, 44(9): 1104-1114. |
| [2] | 施灵玲, 程燕咏, 张磊. 七氟烷对原代少突胶质细胞增殖和分化的影响[J]. 上海交通大学学报(医学版), 2024, 44(9): 1115-1123. |
| [3] | 钱立恒, 温凯玲, 廖颖娜, 李书鑫, 聂惠贞. 分选链接蛋白1抑制结直肠癌细胞增殖和迁移的作用和机制研究[J]. 上海交通大学学报(医学版), 2024, 44(9): 1124-1135. |
| [4] | 何蕊, 颜克鹏, 王静. 靶向髓源性抑制细胞的叶酸循环增强肿瘤免疫治疗效果研究[J]. 上海交通大学学报(医学版), 2024, 44(8): 1011-1022. |
| [5] | 胡飞, 蔡晓涵, 程睿, 季诗雨, 苗嘉欣, 朱晏, 范广建. 骨肉瘤免疫微环境及其免疫治疗临床转化研究进展[J]. 上海交通大学学报(医学版), 2024, 44(7): 814-821. |
| [6] | 张烨晟, 杨易静, 黄依雯, 施珑玙, 王曼媛, 陈思思. 肿瘤微环境免疫细胞调节肿瘤细胞耐药性的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(7): 830-838. |
| [7] | 谭露, 沈少明, 何平. 低氧诱导的长链非编码RNA 68在肝癌中的功能及其作用机制[J]. 上海交通大学学报(医学版), 2024, 44(6): 702-712. |
| [8] | 李萍, 蒋惠如, 叶梦月, 王雅玉, 陈潇雨, 袁安彩, 徐文杰, 戴慧敏, 陈曦, 闫小响, 涂圣贤, 郑元琦, 张薇, 卜军. 基于上海社区老年人群队列的心血管疾病和恶性肿瘤的危险因素流行特征分析[J]. 上海交通大学学报(医学版), 2024, 44(5): 617-625. |
| [9] | 蔡人杰, 徐明. KHSRP通过ANK3调节前列腺癌细胞对雄激素的反应性[J]. 上海交通大学学报(医学版), 2024, 44(4): 417-426. |
| [10] | 安俊伊, 陈必颖, 陈循睿, 尹姗姗, 边洲亮, 刘峰. SFXN3在头颈部鳞状细胞癌中的表达及其对细胞增殖的影响[J]. 上海交通大学学报(医学版), 2024, 44(4): 427-434. |
| [11] | 王梦婷, 陈怡楠, 轩辕欣阳, 袁海花. 肺癌恶性胸腔积液来源肿瘤细胞的小鼠PDX模型构建及实验验证[J]. 上海交通大学学报(医学版), 2024, 44(4): 435-443. |
| [12] | 刘林楠, 冯莉, 王龙, 刘嘉寅, 范志松. 多能蛋白聚糖在恶性肿瘤中的表达及生物学作用的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(4): 525-530. |
| [13] | 徐文晖, 杨畅, 李瑞卿, 卞京, 李夏伊, 郑磊贞. 干扰素调节因子3促结直肠癌细胞增殖与侵袭相关探索[J]. 上海交通大学学报(医学版), 2024, 44(3): 301-311. |
| [14] | 刘美伶, 周亚兵, 王晓强. 儿童神经纤维瘤病1型颅内肿瘤性病变的治疗进展[J]. 上海交通大学学报(医学版), 2024, 44(3): 399-406. |
| [15] | 李虎虓, 李笑甜, 赵旭日, 张桓瑜, 周薇, 宋忠臣. 牙龈素提取物对口腔鳞癌细胞HN6生物学特性的影响[J]. 上海交通大学学报(医学版), 2024, 44(2): 161-168. |
| 阅读次数 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
全文 1670
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
摘要 267
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||