
上海交通大学学报(医学版) ›› 2024, Vol. 44 ›› Issue (7): 839-846.doi: 10.3969/j.issn.1674-8115.2024.07.005
• 转化医学研究前沿进展专题 • 上一篇
夏西茜1( ), 丁珂珂1, 张慧恒1, 彭旭飞1, 孙昳旻1, 唐雅珺1, 汤晓芳2(
), 丁珂珂1, 张慧恒1, 彭旭飞1, 孙昳旻1, 唐雅珺1, 汤晓芳2( )
)
收稿日期:2024-01-30
接受日期:2024-04-03
出版日期:2024-07-28
发布日期:2024-07-28
通讯作者:
汤晓芳
E-mail:xiaxixi0715@163.com;tangxiaofang19840@163.com
作者简介:夏西茜(2000—),女,硕士生;电子信箱:xiaxixi0715@163.com。
基金资助:
XIA Xixi1( ), DING Keke1, ZHANG Huiheng1, PENG Xufei1, SUN Yimin1, TANG Yajun1, TANG Xiaofang2(
), DING Keke1, ZHANG Huiheng1, PENG Xufei1, SUN Yimin1, TANG Yajun1, TANG Xiaofang2( )
)
Received:2024-01-30
Accepted:2024-04-03
Online:2024-07-28
Published:2024-07-28
Contact:
TANG Xiaofang
E-mail:xiaxixi0715@163.com;tangxiaofang19840@163.com
Supported by:摘要:
据估计,全球约有700万人受炎症性肠病(inflammatory bowel disease,IBD)的影响,对医疗系统和社会造成了极大负担。在IBD的发生、进展及治疗过程中,肠道菌群及其关键代谢产物——胆汁酸扮演着至关重要的角色。肠道菌群不仅参与胆汁酸的生物转化,丰富胆汁酸的多样性,还通过法尼酯X受体(farnesoid X receptor,FXR)调控其合成与转运过程。同时,胆汁酸亦通过对微生物多样性的支持、直接毒性、间接抗微生物途径和对微生物代谢能力的影响,参与调整肠道菌群的结构和功能。此外,在正常生理条件下,经肠道菌群修饰后的胆汁酸能够促进肠上皮屏障的损伤修复过程,并且通过调节辅助性T细胞(helper T cell,Th细胞)17、调节性T细胞(regulatory T cell,Treg细胞)、CD8+ T细胞和自然杀伤T细胞(natural killer T cell,NKT细胞)等多种免疫细胞功能,促进免疫系统的平衡,减缓IBD的发展。该文重点探讨了肠道菌群通过介导胆汁酸在IBD的发生和发展中发挥的作用,并探索以肠道菌群和胆汁酸为靶点的新型有效治疗策略,如胆汁酸受体调节剂、益生菌和益生元干预、粪便菌群移植(fecal microbiota transplantation,FMT)以及噬菌体疗法等。
中图分类号:
夏西茜, 丁珂珂, 张慧恒, 彭旭飞, 孙昳旻, 唐雅珺, 汤晓芳. 肠道菌群介导胆汁酸影响炎症性肠病的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(7): 839-846.
XIA Xixi, DING Keke, ZHANG Huiheng, PENG Xufei, SUN Yimin, TANG Yajun, TANG Xiaofang. Research progress of the role of intestinal microbiota-mediated bile acids in inflammatory bowel disease[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(7): 839-846.
| Phylum | Genus (species) | Imbalance in IBD patient |
|---|---|---|
| Proteobacteria | Escherichia (AIEC) | ↑ |
| Bacteroidetes | Bacteroides (ETBF) | ↑ |
| Fusobacteria | Fusobacterium | ↑ |
| Firmicutes | Ruminococcus | ↑ |
| Roseburia | ↓ | |
| Faecalibacterium (Faecalibacterium prausnitzii) | ↓ | |
| Actinobacteria | Bifidobacterium | ↓ |
| Verrucomicrobia | Akkermansia (Akkermansia muciniphila) | ↓ |
表1 IBD患者肠道菌群的特征性变化
Tab 1 Intestinal microbial signatures in patients with IBD
| Phylum | Genus (species) | Imbalance in IBD patient |
|---|---|---|
| Proteobacteria | Escherichia (AIEC) | ↑ |
| Bacteroidetes | Bacteroides (ETBF) | ↑ |
| Fusobacteria | Fusobacterium | ↑ |
| Firmicutes | Ruminococcus | ↑ |
| Roseburia | ↓ | |
| Faecalibacterium (Faecalibacterium prausnitzii) | ↓ | |
| Actinobacteria | Bifidobacterium | ↓ |
| Verrucomicrobia | Akkermansia (Akkermansia muciniphila) | ↓ |
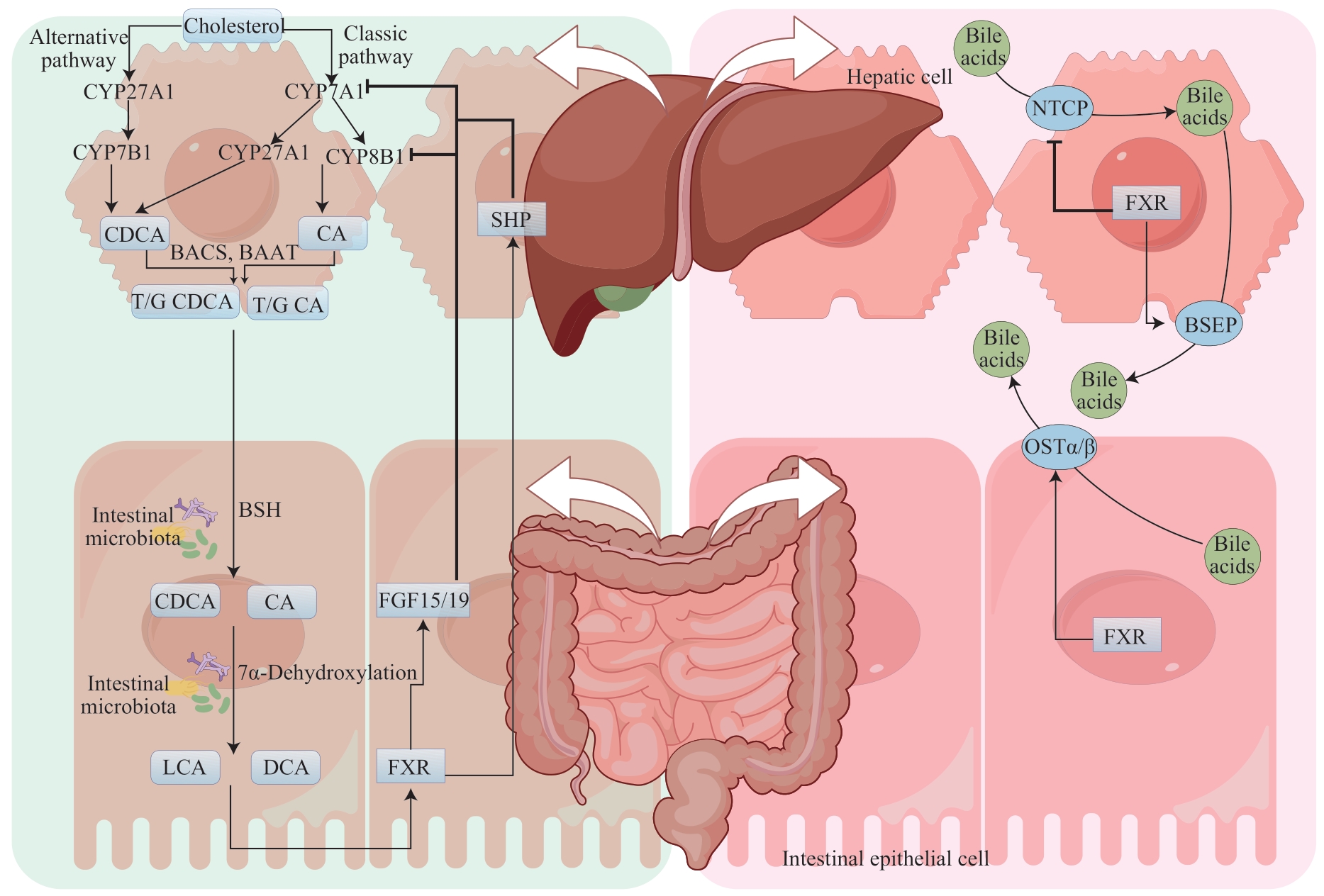
图1 肠道菌群通过调节FXR影响胆汁酸代谢Note: BAAT—bile acid-CoA: amino acid N-acyltransferase; BACS—bile acid coenzyme A synthetase; T/G CDCA—taurochenodeoxycholic/glycochenodeoxycholic acid; T/G CA—taurocholate/ glycocholic acid.
Fig1 Intestinal microbiota affects bile acids metabolism by regulating FXR
| 1 | SHAO B L, YANG W J, CAO Q. Landscape and predictions of inflammatory bowel disease in China: China will enter the Compounding Prevalence stage around 2030[J]. Front Public Health, 2022, 10: 1032679. |
| 2 | KAPLAN G G, WINDSOR J W. The four epidemiological stages in the global evolution of inflammatory bowel disease[J]. Nat Rev Gastroenterol Hepatol, 2021, 18(1): 56-66. |
| 3 | SARTOR R B, WU G D. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches[J]. Gastroenterology, 2017, 152(2): 327-339.e4. |
| 4 | LIU S, ZHAO W J, LAN P, et al. The microbiome in inflammatory bowel diseases: from pathogenesis to therapy[J]. Protein Cell, 2021, 12(5): 331-345. |
| 5 | SHAN Y, LEE M, CHANG E B. The gut microbiome and inflammatory bowel diseases[J]. Annu Rev Med, 2022, 73: 455-468. |
| 6 | QUINN R A, MELNIK A V, VRBANAC A, et al. Global chemical effects of the microbiome include new bile-acid conjugations[J]. Nature, 2020, 579(7797): 123-129. |
| 7 | SONG Z W, CAI Y Y, LAO X Z, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome[J]. Microbiome, 2019, 7(1): 9. |
| 8 | TANG B, TANG L, LI S P, et al. Gut microbiota alters host bile acid metabolism to contribute to intrahepatic cholestasis of pregnancy[J]. Nat Commun, 2023, 14(1): 1305. |
| 9 | GOODWIN B, JONES S A, PRICE R R, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis[J]. Mol Cell, 2000, 6(3): 517-526. |
| 10 | KONG B, WANG L, CHIANG J Y, et al. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice[J]. Hepatology, 2012, 56(3): 1034-1043. |
| 11 | DENSON L A, STURM E, ECHEVARRIA W, et al. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp[J]. Gastroenterology, 2001, 121(1): 140-147. |
| 12 | CHIANG J Y L, FERRELL J M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy[J]. Am J Physiol Gastrointest Liver Physiol, 2020, 318(3): G554-G573. |
| 13 | SUN L L, XIE C, WANG G, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin[J]. Nat Med, 2018, 24(12): 1919-1929. |
| 14 | ZHANG X Q, OSAKA T, TSUNEDA S. Bacterial metabolites directly modulate farnesoid X receptor activity[J]. Nutr Metab, 2015, 12: 48. |
| 15 | VAN BEST N, ROLLE-KAMPCZYK U, SCHAAP F G, et al. Bile acids drive the newborn′s gut microbiota maturation[J]. Nat Commun, 2020, 11(1): 3692. |
| 16 | TIAN Y, GUI W, KOO I, et al. The microbiome modulating activity of bile acids[J]. Gut Microbes, 2020, 11(4): 979-996. |
| 17 | WATANABE M, FUKIYA S, YOKOTA A. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents[J]. J Lipid Res, 2017, 58(6): 1143-1152. |
| 18 | LI Y, TANG R Q, LEUNG P S C, et al. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases[J]. Autoimmun Rev, 2017, 16(9): 885-896. |
| 19 | CREMERS C M, KNOEFLER D, VITVITSKY V, et al. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo[J]. Proc Natl Acad Sci U S A, 2014, 111(16): E1610-E1619. |
| 20 | D'ALDEBERT E, BIYEYEME BI MVE M J, MERGEY M, et al. Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium[J]. Gastroenterology, 2009, 136(4): 1435-1443. |
| 21 | INAGAKI T, MOSCHETTA A, LEE Y K, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor[J]. Proc Natl Acad Sci U S A, 2006, 103(10): 3920-3925. |
| 22 | KAKIYAMA G, PANDAK W M, GILLEVET P M, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis[J]. J Hepatol, 2013, 58(5): 949-955. |
| 23 | MOUSA O Y, JURAN B D, MCCAULEY B M, et al. Bile acid profiles in primary sclerosing cholangitis and their ability to predict hepatic decompensation[J]. Hepatology, 2021, 74(1): 281-295. |
| 24 | SINHA S R, HAILESELASSIE Y, NGUYEN L P, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation[J]. Cell Host Microbe, 2020, 27(4): 659-670.e5. |
| 25 | XU M Q, CEN M S, SHEN Y Q, et al. Deoxycholic acid-induced gut dysbiosis disrupts bile acid enterohepatic circulation and promotes intestinal inflammation[J]. Dig Dis Sci, 2021, 66(2): 568-576. |
| 26 | LI T, DING N, GUO H Q, et al. A gut microbiota-bile acid axis promotes intestinal homeostasis upon aspirin-mediated damage[J]. Cell Host Microbe, 2024, 32(2): 191-208.e9. |
| 27 | CHEN L, JIAO T Y, LIU W W, et al. Hepatic cytochrome P450 8B1 and cholic acid potentiate intestinal epithelial injury in colitis by suppressing intestinal stem cell renewal[J]. Cell Stem Cell, 2022, 29(9): 1366-1381.e9. |
| 28 | JIANG W Y, SU J W, ZHANG X F, et al. Elevated levels of Th17 cells and Th17-related cytokines are associated with disease activity in patients with inflammatory bowel disease[J]. Inflamm Res, 2014, 63(11): 943-950. |
| 29 | PAIK D, YAO L N, ZHANG Y C, et al. Human gut bacteria produce Τh17-modulating bile acid metabolites[J]. Nature, 2022, 603(7903): 907-912. |
| 30 | CARUSO R, LO B C, NÚÑEZ G. Host-microbiota interactions in inflammatory bowel disease[J]. Nat Rev Immunol, 2020, 20(7): 411-426. |
| 31 | SONG X Y, SUN X M, OH S F, et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis[J]. Nature, 2020, 577(7790): 410-415. |
| 32 | LI W, HANG S Y, FANG Y, et al. A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1[J]. Cell Host Microbe, 2021, 29(9): 1366-1377.e9. |
| 33 | LEE J C, LYONS P A, MCKINNEY E F, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis[J]. J Clin Invest, 2011, 121(10): 4170-4179. |
| 34 | DING C J, HONG Y, CHE Y, et al. Bile acid restrained T cell activation explains cholestasis aggravated hepatitis B virus infection[J]. FASEB J, 2022, 36(9): e22468. |
| 35 | ZHU C, BOUCHERON N, MÜLLER A C, et al. 24-Norursodeoxycholic acid reshapes immunometabolism in CD8+ T cells and alleviates hepatic inflammation[J]. J Hepatol, 2021, 75(5): 1164-1176. |
| 36 | KHAN K J, ULLMAN T A, FORD A C, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis[J]. Am J Gastroenterol, 2011, 106(4): 661-673. |
| 37 | HU C L, LIAO S T, LV L, et al. Intestinal immune imbalance is an alarm in the development of IBD[J]. Mediators Inflamm, 2023, 2023: 1073984. |
| 38 | MA C, HAN M J, HEINRICH B, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells[J]. Science, 2018, 360(6391): eaan5931. |
| 39 | CHENG P, WU J W, ZONG G F, et al. Capsaicin shapes gut microbiota and pre-metastatic niche to facilitate cancer metastasis to liver[J]. Pharmacol Res, 2023, 188: 106643. |
| 40 | SHAO J W, GE T T, TANG C L, et al. Synergistic anti-inflammatory effect of gut microbiota and lithocholic acid on liver fibrosis[J]. Inflamm Res, 2022, 71(10/11): 1389-1401. |
| 41 | CHEN Y, LE T H, DU Q M, et al. Genistein protects against DSS-induced colitis by inhibiting NLRP3 inflammasome via TGR5-cAMP signaling[J]. Int Immunopharmacol, 2019, 71: 144-154. |
| 42 | CAMPBELL C, MCKENNEY P T, KONSTANTINOVSKY D, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells[J]. Nature, 2020, 581(7809): 475-479. |
| 43 | FAN L N, QI Y D, QU S W, et al. B. adolescentis ameliorates chronic colitis by regulating Treg/Th2 response and gut microbiota remodeling[J]. Gut Microbes, 2021, 13(1): 1-17. |
| 44 | ALMO M M D, SOUSA I G, OLINTO V G, et al. Therapeutic effects of Zymomonas mobilis on experimental DSS-induced colitis mouse model[J]. Microorganisms, 2023, 11(11): 2793. |
| 45 | ZHOU J, LI M Y, CHEN Q F, et al. Programmable probiotics modulate inflammation and gut microbiota for inflammatory bowel disease treatment after effective oral delivery[J]. Nat Commun, 2022, 13(1): 3432. |
| 46 | VALCHEVA R, KOLEVA P, MARTÍNEZ I, et al. Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels[J]. Gut Microbes, 2019, 10(3): 334-357. |
| 47 | AKRAM W, GARUD N, JOSHI R. Role of inulin as prebiotics on inflammatory bowel disease[J]. Drug Discov Ther, 2019, 13(1): 1-8. |
| 48 | ZHANG Z Z, PAN Y, GUO Z Y, et al. An olsalazine nanoneedle-embedded inulin hydrogel reshapes intestinal homeostasis in inflammatory bowel disease[J]. Bioact Mater, 2024, 33: 71-84. |
| 49 | ARMSTRONG H K, BORDING-JORGENSEN M, SANTER D M, et al. Unfermented β-fructan fibers fuel inflammation in select inflammatory bowel disease patients[J]. Gastroenterology, 2023, 164(2): 228-240. |
| 50 | MOAYYEDI P, SURETTE M G, KIM P T, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial[J]. Gastroenterology, 2015, 149(1): 102-109.e6. |
| 51 | COSTELLO S P, HUGHES P A, WATERS O, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial[J]. JAMA, 2019, 321(2): 156-164. |
| 52 | SOKOL H, LANDMAN C, SEKSIK P, et al. Fecal microbiota transplantation to maintain remission in Crohn′s disease: a pilot randomized controlled study[J]. Microbiome, 2020, 8(1): 12. |
| 53 | KONG L J, LLOYD-PRICE J, VATANEN T, et al. Linking strain engraftment in fecal microbiota transplantation with maintenance of remission in Crohn's disease[J]. Gastroenterology, 2020, 159(6): 2193-2202.e5. |
| 54 | FEDERICI S, KREDO-RUSSO S, VALDÉS-MAS R, et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation[J]. Cell, 2022, 185(16): 2879-2898.e24. |
| 55 | ZHANG L S, WANG Y D, CHEN W D, et al. Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine in mice[J]. Hepatology, 2012, 56(6): 2336-2343. |
| 56 | GADALETA R M, VAN ERPECUM K J, OLDENBURG B, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease[J]. Gut, 2011, 60(4): 463-472. |
| 57 | GOHDA K, IGUCHI Y, MASUDA A, et al. Design and identification of a new farnesoid X receptor (FXR) partial agonist by computational structure-activity relationship analysis: ligand-induced H8 helix fluctuation in the ligand-binding domain of FXR may lead to partial agonism[J]. Bioorg Med Chem Lett, 2021, 41: 128026. |
| [1] | 杜亚格, 卢言慧, 安宇, 宋颖, 郑婕. 肠道菌群在糖尿病认知功能障碍中的作用机制及靶向干预的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(4): 494-500. |
| [2] | 卢雨涵, 石亚红, 龙满美, 王子, 吴颖为. 氧化纳米铈清除活性氧改善DSS诱导的小鼠结肠炎疾病活动度的研究[J]. 上海交通大学学报(医学版), 2024, 44(1): 35-42. |
| [3] | 马锦倩, 范翩翩, 郑涛, 张琳, 陈远志, 申剑, 欧阳凤秀. 孕妇肠道、阴道菌群和新生儿胎粪、胎皮脂菌群的相关性研究[J]. 上海交通大学学报(医学版), 2024, 44(1): 50-63. |
| [4] | 李郡如, 欧阳彦, 谢静远. 肠道菌群在IgA肾病发病与治疗中的作用研究进展[J]. 上海交通大学学报(医学版), 2023, 43(8): 1044-1048. |
| [5] | 高羽, 殷姗, 庞玥, 梁文懿, 刘玉敏. 大黄对大鼠体内肠道菌群-宿主共代谢作用的影响[J]. 上海交通大学学报(医学版), 2023, 43(8): 997-1007. |
| [6] | 温亚锦, 何雯, 韩晓, 张晓波. 不同严重程度支气管哮喘儿童肠道菌群差异的探索性分析[J]. 上海交通大学学报(医学版), 2023, 43(6): 655-664. |
| [7] | 王洁仪, 郑丹, 郑晓皎, 贾伟, 赵爱华. 茶褐素生物学活性及其作用机制的研究进展[J]. 上海交通大学学报(医学版), 2023, 43(6): 768-774. |
| [8] | 刘芊若, 方子晨, 吴宇涵, 钟羡欣, 国沐禾, 贾洁. 肠道菌群及其代谢产物与妊娠期糖尿病相关性的研究进展[J]. 上海交通大学学报(医学版), 2023, 43(5): 641-647. |
| [9] | 王婕, 吴慧, 卢凌鹏, 杨科峰, 祝捷, 周恒益, 姚蝶, 高雅, 冯宇婷, 刘玉红, 贾洁. 妊娠期糖尿病女性肠道菌群的变化特征及其与血糖、血脂和膳食的相关性[J]. 上海交通大学学报(医学版), 2022, 42(9): 1336-1346. |
| [10] | 卢雨, 王昊, 巴乾. 肠道菌群在肝癌发生发展及治疗中的作用研究进展[J]. 上海交通大学学报(医学版), 2022, 42(7): 939-944. |
| [11] | 李博文, 刘宁宁, 王慧. 肠道微生物组在炎症性肠病发病机制和治疗中的作用研究进展[J]. 上海交通大学学报(医学版), 2022, 42(3): 364-368. |
| [12] | 蒋怡, 江平, 张明明, 房静远. 嗜黏蛋白阿克曼菌在肠道相关疾病中作用的研究进展[J]. 上海交通大学学报(医学版), 2022, 42(10): 1490-1497. |
| [13] | 杨紫瑜, 秦娟秀, 李敏, 刘倩. 代谢调控蛋白A调控革兰阳性菌代谢与毒力偶联的研究进展[J]. 上海交通大学学报(医学版), 2021, 41(4): 535-539. |
| [14] | 吴静, 李学义, 陈京红, 王泽剑. 抑郁模型小鼠海马中胆汁酸受体变化的研究[J]. 上海交通大学学报(医学版), 2021, 41(12): 1628-1634. |
| [15] | 袁瑞雪, 傅迎美, 禹顺英. 辅助性T细胞17和调节性T细胞在抑郁症中的作用机制研究进展[J]. 上海交通大学学报(医学版), 2021, 41(10): 1384-1388. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||