
上海交通大学学报(医学版) ›› 2025, Vol. 45 ›› Issue (11): 1443-1457.doi: 10.3969/j.issn.1674-8115.2025.11.004
• 论著 · 基础研究 • 上一篇
张舒琼1,2,3,4, 柯星1, 赵兴贺1, 陈晓翠1, 郑浩东1, 陈惠1, 沈立松1,2,3( ), 杨俊瑶1,3,5(
), 杨俊瑶1,3,5( )
)
收稿日期:2025-05-29
接受日期:2025-06-30
出版日期:2025-11-28
发布日期:2025-12-03
通讯作者:
沈立松,教授,博士;电子信箱:lisongshen@hotmail.com作者简介:第一联系人:为共同第一作者(co-first authors)。
基金资助:
ZHANG Shuqiong1,2,3,4, KE Xing1, ZHAO Xinghe1, CHEN Xiaocui1, ZHENG Haodong1, CHEN Hui1, SHEN Lisong1,2,3( ), YANG Junyao1,3,5(
), YANG Junyao1,3,5( )
)
Received:2025-05-29
Accepted:2025-06-30
Online:2025-11-28
Published:2025-12-03
Contact:
SHEN Lisong, E-mail: lisongshen@hotmail.comSupported by:摘要:
目的·研究长链非编码RNA LINC01123在胃癌中的表达及其促进肿瘤进展的作用机制,探讨LINC01123成为一种新的胃癌诊断标志物和治疗靶点的可能性。方法·基于癌症基因组图谱(The Cancer Genome Atlas,TCGA)和基因表达综合(Gene Expression Omnibus,GEO)数据库(GSE95667、GSE99416)分析胃癌组织与正常组织中lncRNA的表达差异,使用R语言筛选并确定研究对象LINC01123。利用qRT-PCR验证LINC01123在胃癌细胞系及组织样本中的表达水平。通过基因过表达及敲减,利用CCK-8、克隆形成、划痕实验以及Transwell迁移与侵袭实验体外评估LINC01123对细胞增殖和转移能力的影响;构建裸鼠皮下成瘤模型,观察LINC01123对肿瘤生长的促进作用;检测ATP和乳酸浓度以验证LINC01123对糖酵解的调控能力。进一步通过转录物组测序及基因集富集分析(Gene Set Enrichment Analysis,GSEA)探索其可能参与的分子通路。RNA pull-down联合质谱分析初步筛选LINC01123结合蛋白,结合catRAPID数据库预测并通过蛋白质截断突变体和RNA免疫沉淀(RNA immunoprecipitation,RIP)实验确定关键结合区域;通过功能相互作用实验明确α-烯醇化酶(α-enolase,ENO1)是否介导LINC01123的促肿瘤效应。结果·LINC01123在胃癌组织和细胞中显著高表达,并与患者的不良预后密切相关(P=0.021)。功能实验表明,LINC01123显著促进胃癌细胞如MKN-45细胞的增殖、迁移、侵袭和糖酵解,且在裸鼠体内促进肿瘤生长(均P<0.05)。机制研究显示,LINC01123主要定位于细胞质,并可与ENO1蛋白的第97~237位氨基酸区域直接结合。进一步发现,在LINC01123过表达胃癌细胞中联合敲减ENO1,部分逆转LINC01123介导的胃癌细胞如MKN-45的促增殖和促迁移效应(均P<0.05),提示ENO1增加可增强LINC01123的在胃癌进展中的促进作用。结论·LINC01123在胃癌中呈高表达,并通过与ENO1蛋白结合促进细胞增殖、转移和糖酵解,进而促进肿瘤进展。其有望作为胃癌的新型诊断和预后生物标志物及治疗靶点。
中图分类号:
张舒琼, 柯星, 赵兴贺, 陈晓翠, 郑浩东, 陈惠, 沈立松, 杨俊瑶. LINC01123通过结合ENO1促进胃癌的增殖和糖酵解[J]. 上海交通大学学报(医学版), 2025, 45(11): 1443-1457.
ZHANG Shuqiong, KE Xing, ZHAO Xinghe, CHEN Xiaocui, ZHENG Haodong, CHEN Hui, SHEN Lisong, YANG Junyao. LINC01123 promotes proliferation and glycolysis of gastric cancer via binding to ENO1[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(11): 1443-1457.
| Gene | Sequence (5′→3′) |
|---|---|
| si-LINC01123 | |
| Sense | TTCTCTTTATTTTTATACAGT |
| Antisense | ACTGTATAAAAATAAAGAGAA |
| si-ENO1 | |
| Sense | CCCAGUGGUGUCUAUCGAATT |
| Antisense | UUCGAUAGACACCACUGGGTT |
| si-NC | |
| Sense | UUCUCCGAACGUGUCACGU |
| Antisense | ACGUGACACGUUCGGAGAA |
| sh-LINC01123 | CCGGTTCTCTTTATTTTTATACAGTCTCGAGACTGTATAAAAATAAAGAGAATTTTTG |
| sh-NC | TTCTCCGAACGTGTCACGT |
表1 慢病毒载体构建使用的敲减序列
Tab 1 Knockdown sequences for lentiviral vector construction
| Gene | Sequence (5′→3′) |
|---|---|
| si-LINC01123 | |
| Sense | TTCTCTTTATTTTTATACAGT |
| Antisense | ACTGTATAAAAATAAAGAGAA |
| si-ENO1 | |
| Sense | CCCAGUGGUGUCUAUCGAATT |
| Antisense | UUCGAUAGACACCACUGGGTT |
| si-NC | |
| Sense | UUCUCCGAACGUGUCACGU |
| Antisense | ACGUGACACGUUCGGAGAA |
| sh-LINC01123 | CCGGTTCTCTTTATTTTTATACAGTCTCGAGACTGTATAAAAATAAAGAGAATTTTTG |
| sh-NC | TTCTCCGAACGTGTCACGT |
| Gene | Sequence (5′→3′) |
|---|---|
| LINC01123 | |
| Forward | ACAGTGGCCGCACGCATAGCTG |
| Reverse | CTGACGACCGAGGTGACAACGATGA |
| ENO1 | |
| Forward | GCCTCCTGCTCAAAGTCAAC |
| Reverse | AACGATGAGACACCATGACG |
| GAPDH | |
| Forward | TTGGTATCGTGGAAGGACTCA |
| Reverse | TGTCATCATATTTGGCAGGTTT |
| U6 | |
| Forward | CGCTTCGGCAGCACATATAC |
| Reverse | TTCACGAATTTGCGTGTCATC |
表2 qRT-PCR引物序列
Tab 2 Primers sequences for qRT-PCR
| Gene | Sequence (5′→3′) |
|---|---|
| LINC01123 | |
| Forward | ACAGTGGCCGCACGCATAGCTG |
| Reverse | CTGACGACCGAGGTGACAACGATGA |
| ENO1 | |
| Forward | GCCTCCTGCTCAAAGTCAAC |
| Reverse | AACGATGAGACACCATGACG |
| GAPDH | |
| Forward | TTGGTATCGTGGAAGGACTCA |
| Reverse | TGTCATCATATTTGGCAGGTTT |
| U6 | |
| Forward | CGCTTCGGCAGCACATATAC |
| Reverse | TTCACGAATTTGCGTGTCATC |
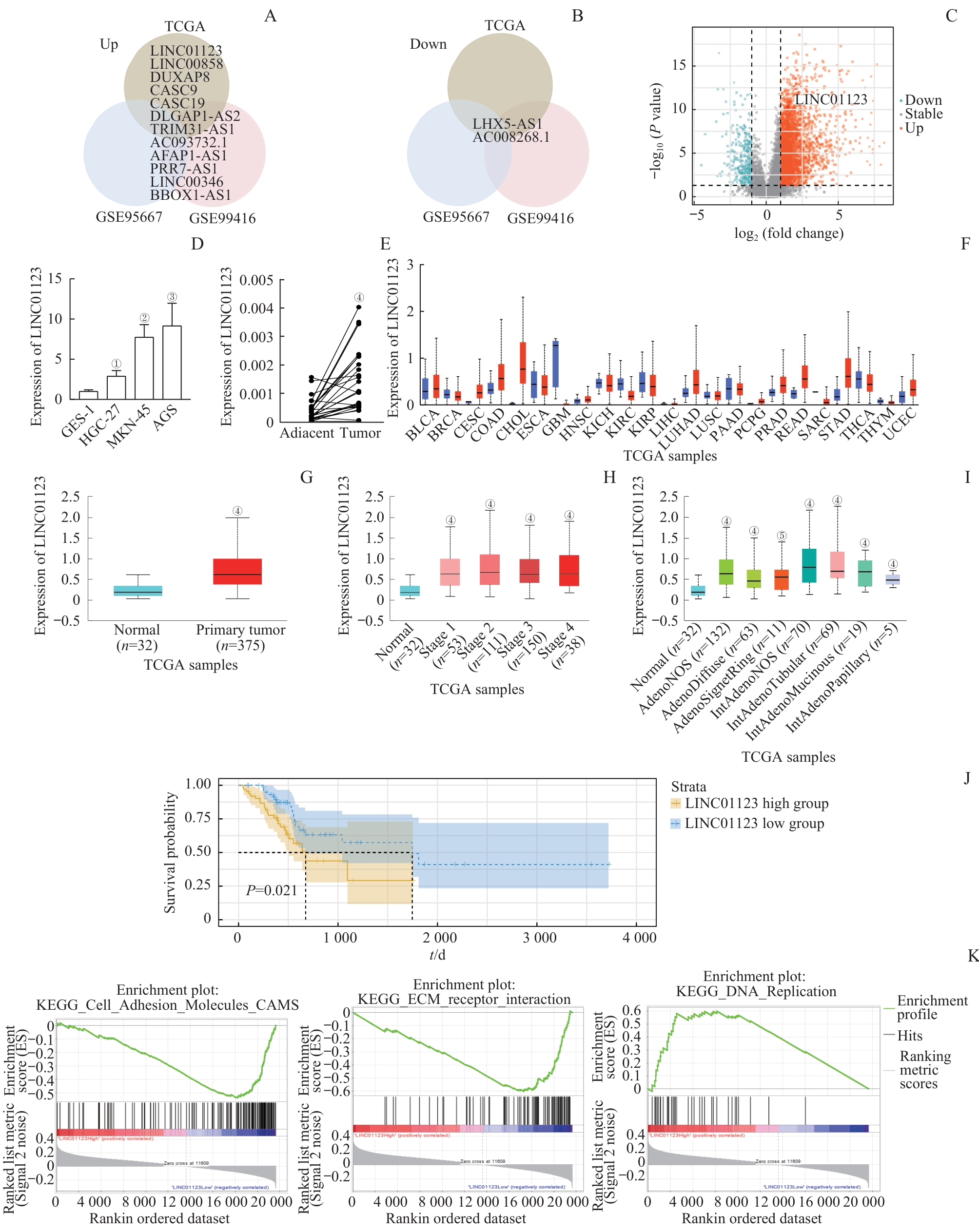
图1 LINC01123在胃癌中的高表达及不良预后关联Note: A/B. Venn diagrams illustrating the overlap of upregulated (A) and downregulated (B) long non-coding RNAs (lncRNAs) identified in the TCGA-STAD, GSE95667, and GSE99416 datasets. C. Volcano plot depicting differentially expressed lncRNAs in the TCGA-STAD dataset. D. LINC01123 expression was significantly upregulated in gastric cancer cell lines. E. LINC01123 expression was significantly upregulated in gastric cancer tissues compared to adjacent normal tissues. F. Expression of LINC01123 across different cancer types in TCGA. G. UALCAN analysis showing that LINC01123 was upregulated in STAD tissues. H. LINC01123 expression in different TNM stages of gastric cancer. I. LINC01123 expression profile based on tumor histology. J. Kaplan-Meier analysis showing that high LINC01123 expression was associated with poor overall survival in TCGA-STAD patients. K. Gene set enrichment analysis (GSEA) comparing high and low LINC01123 expression groups. ①P=0.011, ②P=0.002, ③P=0.007, compared with GES-1; ④P<0.001, ⑤P=0.004, compared with Normal.
Fig 1 Overexpression of LINC01123 in gastric cancer and its association with poor prognosis
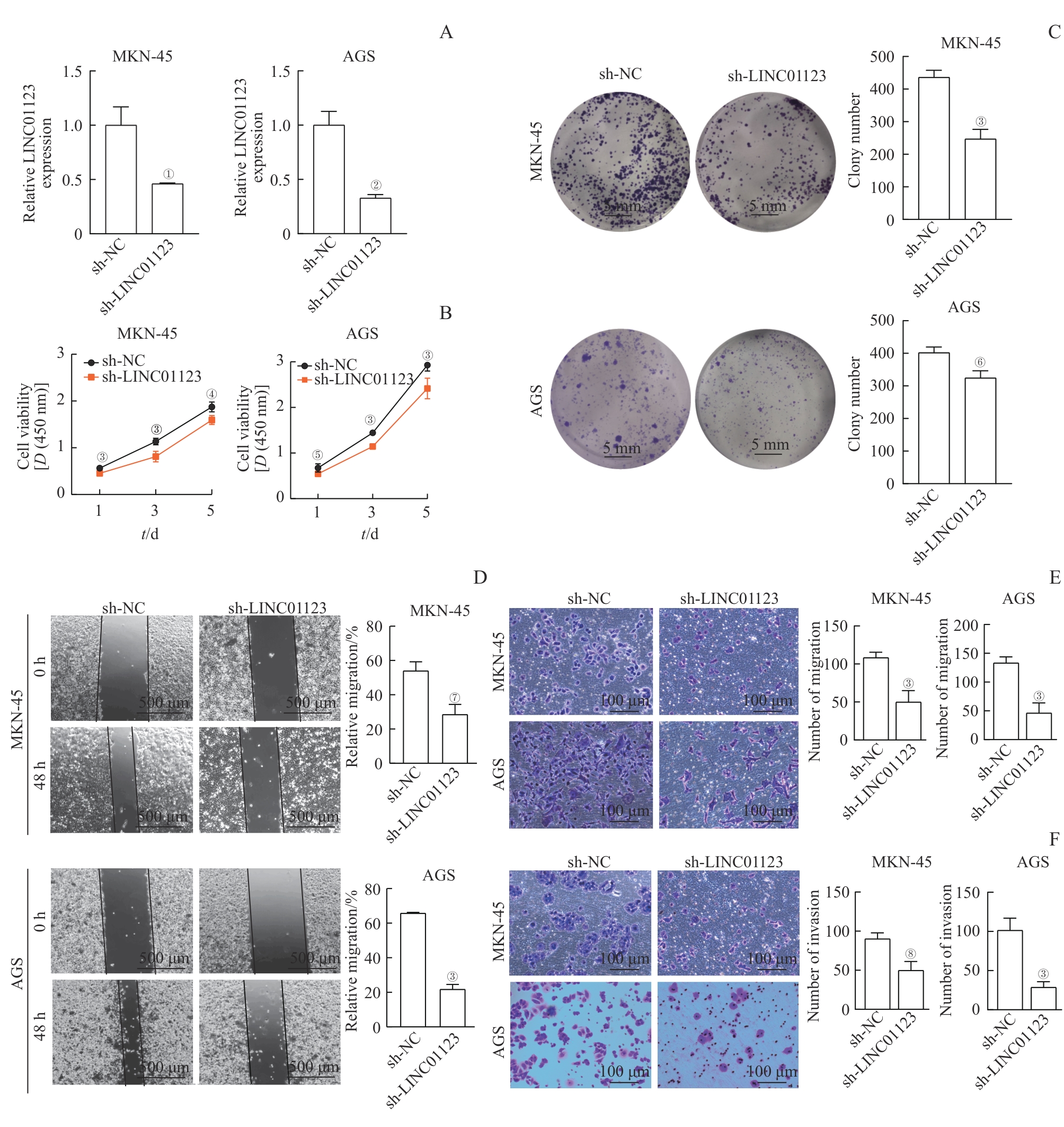
图2 LINC01123敲低对胃癌细胞体外增殖、迁移和侵袭的抑制作用Note: A. qRT-PCR assay demonstrated that sh-LINC01123 lentivirus effectively altered LINC01123 expression. B/C. CCK-8 assay (B) and colony-formation assay (C) showed the effects of knockdown of LINC01123 on cell proliferation. D/E. Wound healing assay (D) and Transwell migration assay (E) were performed to detect the migration ability of MKN-45 and AGS cells after knockdown of LINC01123. F. Transwell invasion assay was conducted to assess the effects of LINC01123 knockdown on cell invasion. ①P=0.030, ②P=0.011, ③P<0.001, ④P=0.010, ⑤P=0.020, ⑥P=0.009, ⑦P=0.005, ⑧P=0.007, compared with sh-NC.
Fig 2 Inhibitory effects of LINC01123 knockdown on gastric cancer cell proliferation, migration, and invasion in vitro
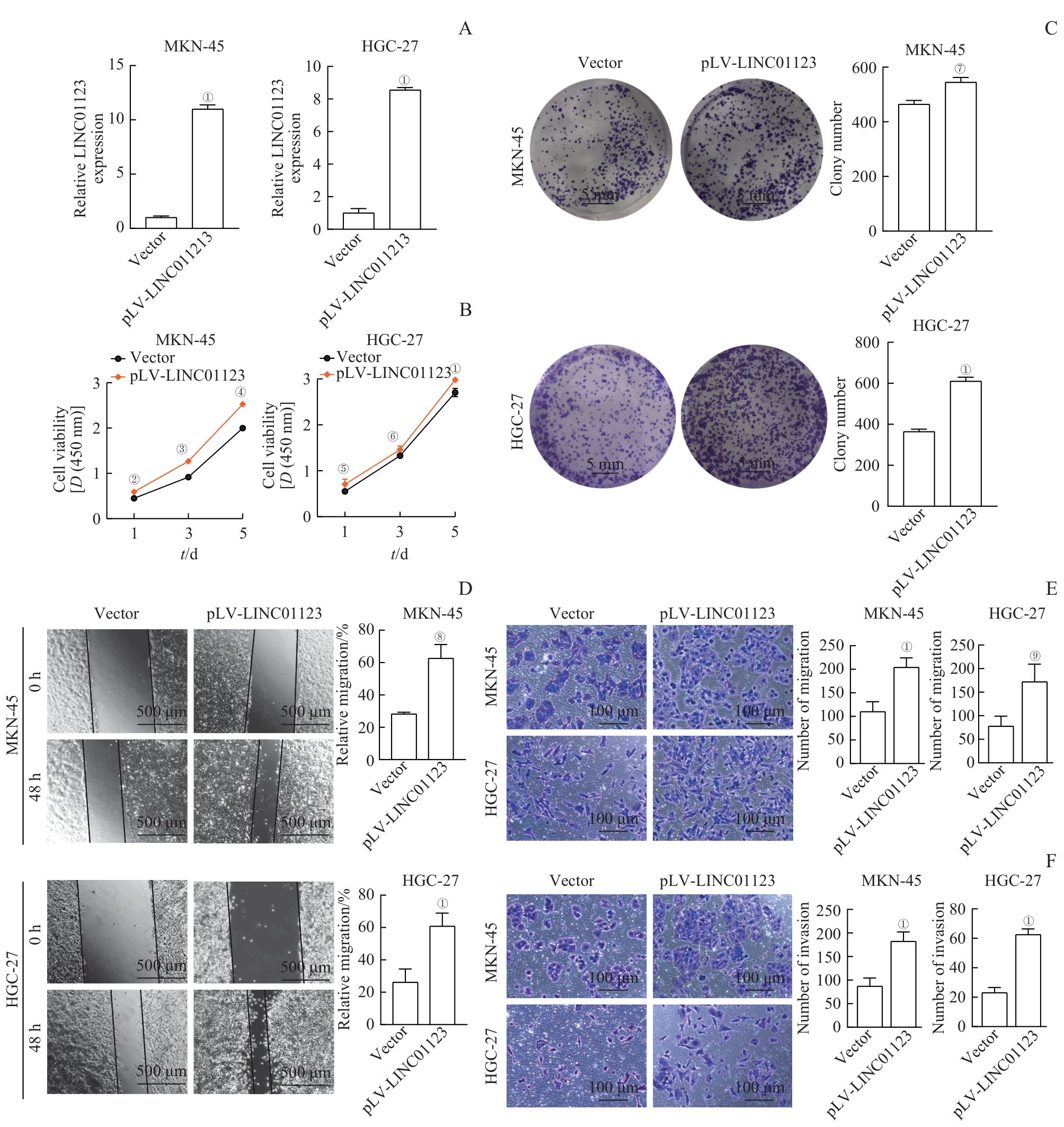
图3 LINC01123过表达对胃癌细胞体外增殖、迁移和侵袭的促进作用Note: A. qRT-PCR assay demonstrated that LINC01123 overexpression lentivirus effectively altered LINC01123 expression. B/C. CCK-8 assay (B) and colony-formation assay (C) showed the effects of overexpression of LINC01123 on cell proliferation. D. Wound healing assay was performed to detect the migration ability of MKN-45 and HGC-27 cells after the overexpression of LINC01123. E. Transwell migration assay was performed to detect the migration ability of MKN-45 and HGC-27 cells after the overexpression of LINC01123. F. Transwell invasion assay was conducted to assess the effects of knockdown or overexpression of LINC01123 on cell invasion. ①P<0.001, ②P=0.006, ③P=0.007, ④P=0.010, ⑤P=0.020, ⑥P=0.009, ⑦P=0.030, ⑧P=0.002, ⑨P=0.005, compared with Vector.
Fig 3 Promotive effects of LINC01123 overexpression on gastric cancer cell proliferation, migration, and invasion in vitro
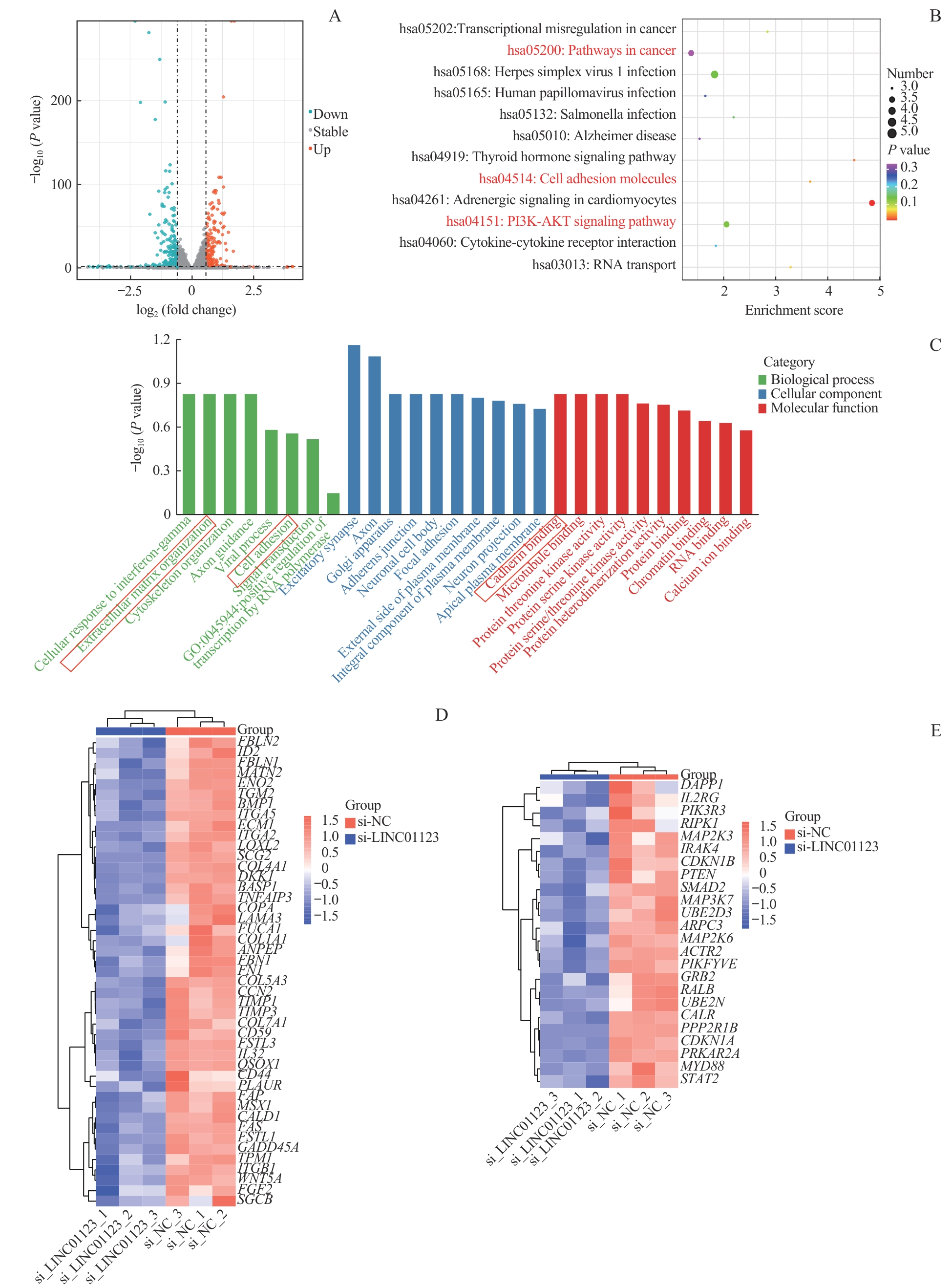
图4 LINC01123对EMT及PI3K/AKT信号通路的调控作用Note:A. Volcano plot of differentially expressed lncRNAs. B. KEGG enrichment results of differentially expressed genes in MKN-45 cells transfected with si-NC and si-LINC01123. C. GO enrichment results of differentially expressed genes in MKN-45 cells transfected with si-NC and si-LINC01123. D. Heatmap of genes defining epithelial-mesenchymal transition (EMT). E. Heatmap of genes up-regulated by activation of the PI3K/AKT/mTOR pathway.
Fig 4 Regulatory role of LINC01123 in EMT and PI3K/AKT pathway
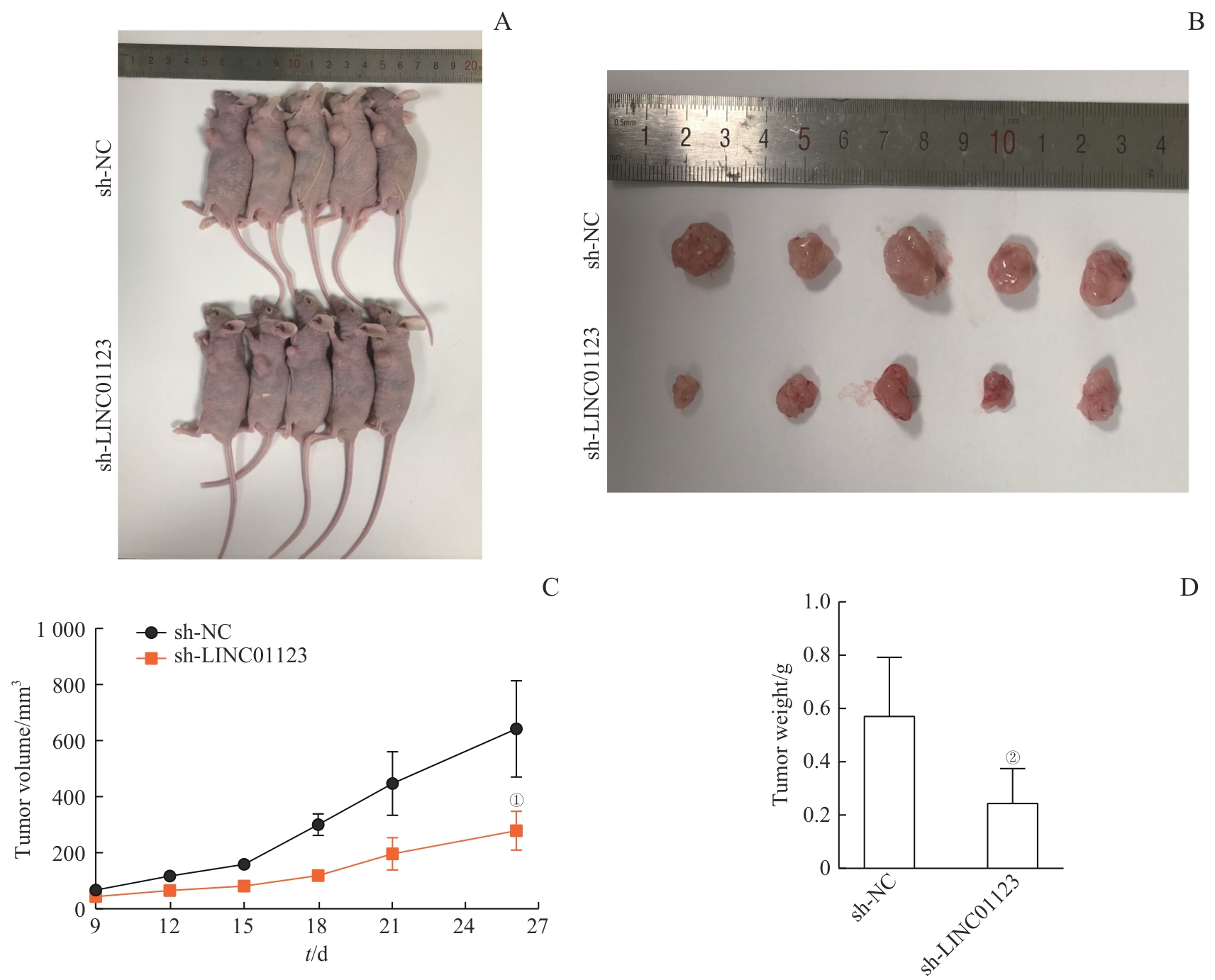
图5 LINC01123在体内促进胃癌MKN-45细胞的成瘤能力Note: A. Representative images of nude mice subcutaneously injected with MKN-45 cells stably expressing sh-NC or sh-LINC01123 lentivirus (n=5 per group). B. Excised tumors from nude mice in sh-NC and sh-LINC01123 groups. C. Tumor volumes of two groups were assessed for 26 d. D. Tumor weight of two groups. ①P<0.001, ②P=0.002, compared with sh-NC.
Fig 5 Promotion of tumorigenesis of gastric cancer MKN-45 cells by LINC01123 in vivo
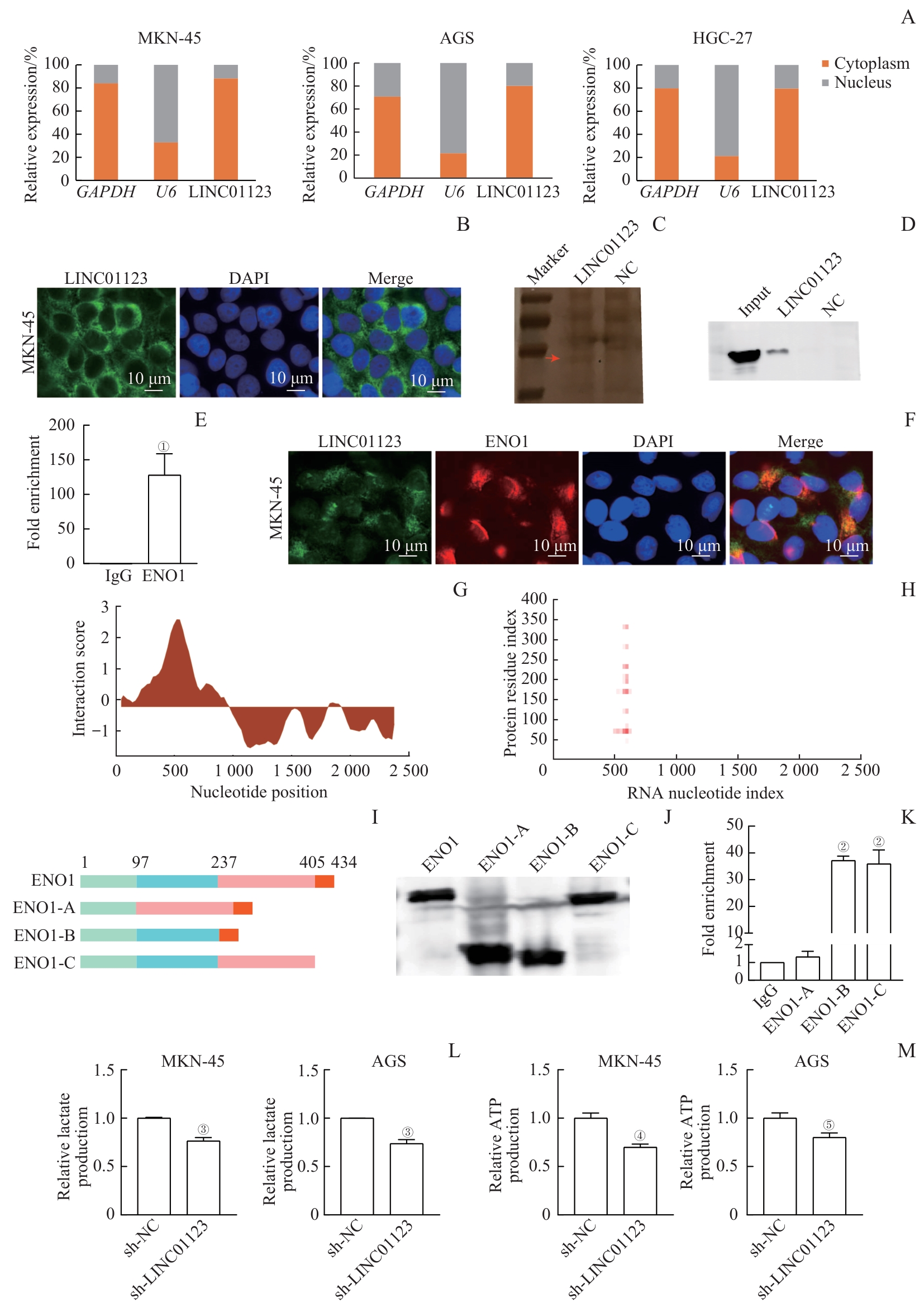
图6 LINC01123与ENO1蛋白的结合及对胃癌细胞糖酵解的调节作用Note: A. Expression of LINC01123 in the cytoplasm and nucleus of gastric cancer cells. B. Fluorescence in situ hybridization (FISH) assay showing the localization of LINC01123 in MKN-45 cells. C. Silver-stained SDS-PAGE of proteins from RNA pull-down assay of MKN-45 cell extracts. The red arrow indicates the band used for mass spectrometry analysis. D. ENO1 pulled down by LINC01123 and negative control RNA was analyzed by Western blotting. Input represents the total protein used for RNA pull-down. E. PCR analysis of LINC01123 expression from RNA immunoprecipitation (RIP) assays. F. Co-staining of LINC01123 (green) and ENO1 (red) in MKN-45 cells by in FISH. G. Schematic map of the potential binding site of LINC01123 in ENO1 by catRAPID analysis. H. The interaction matrix between LINC01123 and ENO1 by catRAPID fragments analysis. I. Graphic display of ENO1 and its truncations. J. Flag-tagged ENO1 and its truncations were verified by Western blotting. K. Quantitative PCR of LINC01123 from purified RNAs of RIP experiments performed with 293T cell extracts transfected with Flag-tagged ENO1 or its truncations. L. Reduced lactate production in MKN-45 and AGS cells after LINC01123 knockdown. M. Reduced ATP production in MKN-45 and AGS cells after LINC01123 knockdown. ①P=0.002, ②P<0.001, compared with IgG; ③P<0.001, ④P=0.009, ⑤P=0.001, compared with sh-NC.
Fig 6 Binding of LINC01123 to ENO1 protein and regulation of glycolysis in gastric cancer cells by LINC01123
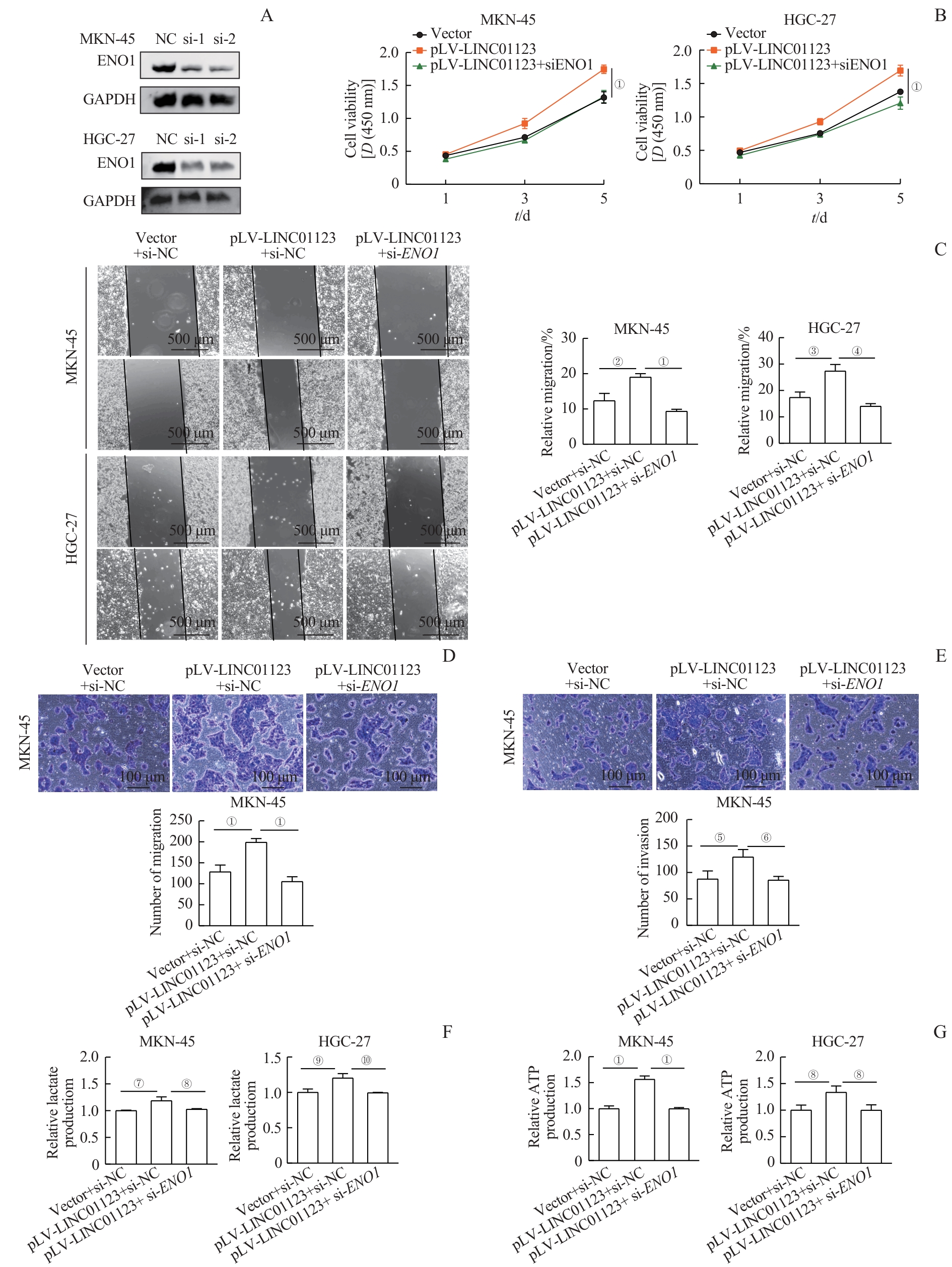
图7 LINC01123通过ENO1发挥的促增殖、迁移、侵袭和糖酵解功能Note: A. ENO1 expression after transfection of ENO1 siRNA in LINC01123-overexpressing gastric cancer MKN-45 and HGC-27 cell lines. B. CCK-8 assay showing that the proliferation of MKN-45 and HGC-27 cells overexpressing LINC01123 decreased after ENO1 knockdown. C. Wound healing assay showing that the migration of MKN-45 and HGC-27 cells overexpressing LINC01123 decreased after ENO1 knockdown. D. Transwell migration assay showing that the migration of MKN-45 cells overexpressing LINC01123 decreased after interference with ENO1 expression. E. Transwell invasion assay showing that the invasion of MKN-45 cells overexpressing LINC01123 decreased after ENO1 knockdown. F/G. ENO1 knockdown reduces LINC01123-induced lactate production (F) and ATP production (G) in MKN-45 and HGC-27 cells overexpressing LINC01123. ①P<0.001, ②P=0.008, ③P=0.006, ④P=0.001, ⑤P=0.007, ⑥P=0.002, ⑦P=0.012, ⑧P=0.020, ⑨P=0.011, ⑩P=0.004.
Fig 7 Promotion of proliferation, migration, invasion, and glycolysis by LINC01123 via ENO1
| [1] | ARNOLD M, PARK J Y, CAMARGO M C, et al. Is gastric cancer becoming a rare disease? A global assessment of predicted incidence trends to 2035[J]. Gut, 2020, 69(5): 823-829. |
| [2] | KIM H, HWANG Y, SUNG H, et al. Effectiveness of gastric cancer screening on gastric cancer incidence and mortality in a community-based prospective cohort[J]. Cancer Res Treat, 2018, 50(2): 582-589. |
| [3] | SIEGEL R L, MILLER K D, WAGLE N S, et al. Cancer statistics, 2023[J]. CA A Cancer J Clinicians, 2023, 73(1): 17-48. |
| [4] | SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249. |
| [5] | PICARD M. Why do we care more about disease than health?[J]. Phenomics, 2022, 2(3): 145-155. |
| [6] | GUTSCHNER T, DIEDERICHS S. The hallmarks of cancer: a long non-coding RNA point of view[J]. RNA Biol, 2012, 9(6): 703-719. |
| [7] | DE LOS SANTOS M C, DRAGOMIR M P, CALIN G A. The role of exosomal long non-coding RNAs in cancer drug resistance[J]. Cancer Drug Resist, 2019, 2(4): 1178-1192. |
| [8] | ZHAO J, XU H, SU Y H, et al. Emerging regulatory mechanisms of N6-methyladenosine modification in cancer metastasis[J]. Phenomics, 2023, 3(1): 83-100. |
| [9] | HUA Q, JIN M M, MI B M, et al. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis[J]. J Hematol Oncol, 2019, 12(1): 91. |
| [10] | SHANG T, ZHOU X, CHEN W. LINC01123 promotes the progression of colorectal cancer via miR-625-5p/LASP1 axis[J]. Cancer Biother Radiopharm, 2021, 36(9): 765-773. |
| [11] | XIAO Z Q, LIU Y, ZHAO J J, et al. Long noncoding RNA LINC01123 promotes the proliferation and invasion of hepatocellular carcinoma cells by modulating the miR-34a-5p/TUFT1 axis[J]. Int J Biol Sci, 2020, 16(13): 2296-2305. |
| [12] | YANG Y, ZHENG Y, ZHANG H L, et al. An immune-related gene panel for preoperative lymph node status evaluation in advanced gastric cancer[J]. Biomed Res Int, 2020, 2020: 8450656. |
| [13] | YE S C, SUN B L, WU W Y, et al. LINC01123 facilitates proliferation, invasion and chemoresistance of colon cancer cells[J]. Biosci Rep, 2020, 40(8): BSR20194062. |
| [14] | ZHANG M, HAN Y G, ZHENG Y, et al. ZEB1-activated LINC01123 accelerates the malignancy in lung adenocarcinoma through NOTCH signaling pathway[J]. Cell Death Dis, 2020, 11(11): 981. |
| [15] | ZHANG P R, LONG Q M, ZENG S Y, et al. FOXC1-induced LINC01123 acts as a mediator in triple negative breast cancer[J]. Cancer Cell Int, 2020, 20: 199. |
| [16] | SUGAHARA T, NAKAJIMA H, SHIRAHATA S, et al. Purification and characterization of immunoglobulin production stimulating factor-II beta derived from Namalwa cells[J]. Cytotechnology, 1992, 10(2): 137-146. |
| [17] | HUANG H, TANG S, JI M, et al. p300-mediated lysine 2-hydroxyisobutyrylation regulates glycolysis[J]. Mol Cell, 2018, 70(4): 663-678.e6. |
| [18] | LÓPEZ-ALEMANY R, LONGSTAFF C, HAWLEY S, et al. Inhibition of cell surface mediated plasminogen activation by a monoclonal antibody against α-enolase[J]. Am J Hematol, 2003, 72(4): 234-242. |
| [19] | HUA Q, WANG D L, ZHAO L, et al. AL355338 acts as an oncogenic lncRNA by interacting with protein ENO1 to regulate EGFR/AKT pathway in NSCLC[J]. Cancer Cell Int, 2021, 21(1): 525. |
| [20] | YU S M, LI N, HUANG Z B, et al. A novel lncRNA, TCONS_00006195, represses hepatocellular carcinoma progression by inhibiting enzymatic activity of ENO1[J]. Cell Death Dis, 2018, 9(12): 1184. |
| [21] | BARLEBO AHLBORN L, ØSTRUP O. Toward liquid biopsies in cancer treatment: application of circulating tumor DNA[J]. APMIS, 2019, 127(5): 329-336. |
| [22] | TRUJANO-CAMACHO S, CANTÚ-DE LEÓN D, PÉREZ-YEPEZ E, et al. HOTAIR promotes the hyperactivation of PI3K/Akt and Wnt/β-catenin signaling pathways via PTEN hypermethylation in cervical cancer[J]. Cells, 2024, 13(17): 1484. |
| [23] | HU Y R, YU Y C, YOU S W, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer[J]. Mol Cancer, 2017, 16(1): 174. |
| [24] | LI H, MA X H, YANG D S, et al. PCAT-1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2[J]. J Cell Biochem, 2020, 121(2): 1353-1361. |
| [25] | GUO Y M, YUE P R, WANG Y M, et al. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis[J]. Biomed Pharmacother, 2019, 118: 109255. |
| [26] | RAY R, MILLER D M. Cloning and characterization of a human c-myc promoter-binding protein[J]. Mol Cell Biol, 1991, 11(4): 2154-2161. |
| [1] | 那迪娜·帕尔哈提, 张鹏善, 徐亦天, 陈赟琪, 黄陈. 人去泛素化酶圆柱瘤蛋白截短体质粒的构建及其对胃癌细胞表型的调控研究[J]. 上海交通大学学报(医学版), 2025, 45(9): 1149-1160. |
| [2] | 朱子俊, 钱逸斐, 李倩玉, 李松玲, 覃雯莉, 刘艳丰. 后期促进复合体亚基10调控PI3K-AKT-mTOR通路促进肝细胞癌进展的研究[J]. 上海交通大学学报(医学版), 2025, 45(9): 1171-1182. |
| [3] | 杨娜, 刘俊丽, 白静, 杨思怡, 韩继明, 张华华. HENMT1通过激活PI3K-AKT-mTOR信号通路促进胃癌的增殖与迁移[J]. 上海交通大学学报(医学版), 2025, 45(6): 717-726. |
| [4] | 许沐馨, 刘贤, 蒋立姗, 孙青. Nd:YAP激光生物刺激通过WNT/β-catenin信号通路促进人牙周韧带细胞增殖和成骨分化[J]. 上海交通大学学报(医学版), 2025, 45(5): 562-569. |
| [5] | 陈怡楠, 郑旸, 曾汉林, 雷鸣. Fas相关死亡结构域蛋白促进头颈部鳞状细胞癌细胞增殖能力的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(4): 404-414. |
| [6] | 陈勇羽, 黄益仁, 陈哲逸, 周冰倩, 陈诗宇, 郑英霞. 丝氨酸蛋白酶抑制因子1在胃癌中的表达及其促进胃癌发展的作用机制[J]. 上海交通大学学报(医学版), 2025, 45(2): 150-160. |
| [7] | 张先洲, 杜凤麟, 吴雷, 任逸喆, 赵明娜, 娄加陶. OGT通过ERK信号通路促进非小细胞肺癌增殖的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(10): 1288-1297. |
| [8] | 李想, 魏鸣, 吴文曦, 罗小琴, 姚彪, 伍思宇. 芦丁对骨肉瘤生长和转移的体内外抑制作用[J]. 上海交通大学学报(医学版), 2025, 45(1): 20-28. |
| [9] | 孙晨玮, 海汪溪, 屈骞, 席云. [18F]F-FMISO和[18F]F-FLT PET/CT双核素显像预测胰腺癌耐药性的体内研究[J]. 上海交通大学学报(医学版), 2025, 45(1): 60-68. |
| [10] | 施灵玲, 程燕咏, 张磊. 七氟烷对原代少突胶质细胞增殖和分化的影响[J]. 上海交通大学学报(医学版), 2024, 44(9): 1115-1123. |
| [11] | 钱立恒, 温凯玲, 廖颖娜, 李书鑫, 聂惠贞. 分选链接蛋白1抑制结直肠癌细胞增殖和迁移的作用和机制研究[J]. 上海交通大学学报(医学版), 2024, 44(9): 1124-1135. |
| [12] | 韩依杉, 徐梓淇, 陶梦玉, 范广建, 余波. PRMT6促进乳腺癌细胞的增殖和迁移[J]. 上海交通大学学报(医学版), 2024, 44(8): 999-1010. |
| [13] | 谭露, 沈少明, 何平. 低氧诱导的长链非编码RNA 68在肝癌中的功能及其作用机制[J]. 上海交通大学学报(医学版), 2024, 44(6): 702-712. |
| [14] | 蔡人杰, 徐明. KHSRP通过ANK3调节前列腺癌细胞对雄激素的反应性[J]. 上海交通大学学报(医学版), 2024, 44(4): 417-426. |
| [15] | 安俊伊, 陈必颖, 陈循睿, 尹姗姗, 边洲亮, 刘峰. SFXN3在头颈部鳞状细胞癌中的表达及其对细胞增殖的影响[J]. 上海交通大学学报(医学版), 2024, 44(4): 427-434. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||