
上海交通大学学报(医学版) ›› 2024, Vol. 44 ›› Issue (12): 1514-1525.doi: 10.3969/j.issn.1674-8115.2024.12.004
刘晨茜1,2( ), 韩林3(
), 韩林3( ), 杨轶1,2, 周韩1,2, 刘亚云1,2(
), 杨轶1,2, 周韩1,2, 刘亚云1,2( ), 盛德乔1,2(
), 盛德乔1,2( )
)
收稿日期:2024-02-26
接受日期:2024-08-05
出版日期:2024-12-28
发布日期:2024-12-28
通讯作者:
刘亚云,电子信箱:liuyyctgu@ctgu.edu.cn。作者简介:刘晨茜(1995—),女,硕士生;电子信箱:381212026@qq.com基金资助:
LIU Chenxi1,2( ), HAN Lin3(
), HAN Lin3( ), YANG Yi1,2, ZHOU Han1,2, LIU Yayun1,2(
), YANG Yi1,2, ZHOU Han1,2, LIU Yayun1,2( ), SHENG Deqiao1,2(
), SHENG Deqiao1,2( )
)
Received:2024-02-26
Accepted:2024-08-05
Online:2024-12-28
Published:2024-12-28
Contact:
LIU Yayun, E-mail: liuyyctgu@ctgu.edu.cn.Supported by:摘要:
目的·探究GPR87在调节非小细胞肺癌(non-small cell lung cancer,NSCLC)侵袭和迁移中的作用及分子机制。方法·利用生物信息学方法,包括GEO、UALCAN、KM Plotter等多个公共数据库分析平台,筛选与NSCLC侵袭相关的候选基因,并预测基因与NSCLC的临床相关性。收集宜昌市中心人民医院2018年1月—2020年8月收治的80例NSCLC临床样本及对应的临床病理资料,利用免疫组化分析肿瘤组织中GPR87的表达,并对GPR87的临床相关性进行分析。用siRNA-GPR87和pCMV-GPR87-his分别转染人肺腺癌细胞系A549和人肺鳞状细胞癌细胞系SK-MES-1,构建低表达和高表达GPR87的细胞系,运用Transwell实验探究GPR87的表达对NSCLC细胞的迁移、侵袭能力的影响,通过酶联免疫吸附试验(enzyme-linked immunosorbent assay,ELISA)检测细胞培养上清液中MMP7的分泌量,用RT-qPCR检测GPR87、MMP2、MMP7、MMP9、E-cadherin、N-cadherin、vimentin、snail、twist、RHOA、RHOC、ROCK1的mRNA表达水平,用ELISA检测MMP7的蛋白分泌量,用Western blotting检测GPR87、MMP9、E-cadherin、vimentin、RHOA、ROCK1的蛋白表达水平。结果·生信分析和临床样本的数据显示,GPR87 mRNA和蛋白在NSCLC中高表达,且会导致患者临床分期更差、更易发生淋巴结转移,提示GPR87可能是NSCLC高侵袭性的关键基因。GPR87表达下调可显著降低A549和SK-MES-1细胞的侵袭和迁移能力,而过表达GPR87可增强A549和SK-MES-1细胞的侵袭和迁移能力。进一步检测发现,GPR87的下调导致A549和SK-MES-1细胞中MMP2、MMP7、MMP9、RHOA、RHOC和ROCK1的mRNA表达水平,MMP7的蛋白分泌量,MMP9、RHOA、ROCK1的蛋白表达水平降低;过表达GPR87增加了细胞MMP2、MMP7、MMP9、RHOA、RHOC和ROCK1的mRNA表达水平、MMP7的蛋白分泌量、MMP9、RHOA、ROCK1的蛋白表达水平。无论GPR87敲低或者过表达,上皮间质转化相关基因和蛋白的表达均无明显变化。结论·GPR87的高表达与NSCLC的高侵袭性密切相关。在SK-MES-1和A549细胞中,GPR87可通过激活RHOA/ROCK1信号通路,促进MMPs的表达,最终促进NSCLC的侵袭与迁移。
中图分类号:
刘晨茜, 韩林, 杨轶, 周韩, 刘亚云, 盛德乔. GPR87通过激活RHO/ROCK通路促进非小细胞肺癌的侵袭和迁移[J]. 上海交通大学学报(医学版), 2024, 44(12): 1514-1525.
LIU Chenxi, HAN Lin, YANG Yi, ZHOU Han, LIU Yayun, SHENG Deqiao. GPR87 promotes invasion and migration through the RHO/ROCK pathway in non-small cell lung cancer[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(12): 1514-1525.
| Name | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| siRNA-GPR87-1 | GACCUUAGUUUCAAAGCUUdTdT | AAGCUUUGAAACUAAGGUCdTdT |
| siRNA-GPR87-2 | GCAUCUUGCUGAAUGGUUUdTdT | AAACCAUUCAGCAAGAUGCdTdT |
| NC-siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
表1 siRNA序列
Tab 1 Sequences of siRNA
| Name | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| siRNA-GPR87-1 | GACCUUAGUUUCAAAGCUUdTdT | AAGCUUUGAAACUAAGGUCdTdT |
| siRNA-GPR87-2 | GCAUCUUGCUGAAUGGUUUdTdT | AAACCAUUCAGCAAGAUGCdTdT |
| NC-siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| GPR87 | GGAGGCGACATCAATGCAG | AAATGAAAGTAAAGAACGATTTTGTGT |
| MMP2 | GCTGGAGACAAATTCTGGAGATACA | GTATCGAAGGCAGTGGAGAGGA |
| MMP7 | GAGTGAGCTACAGTGGGAACA | CTATGACGCGGGAGTTTAACAT |
| MMP9 | GCCACTACTGTGCCTTTGAGTC | CCCTCAGAGAATCGCCAGTACT |
| E-cadherin | GCCTCCTGAAAAGAGAGTGGAAG | TGGCAGTGTCTCTCCAAATCCG |
| N-cadherin | CCTCCAGAGTTTACTGCCATGAC | GTAGGATCTCCGCCACTGATTC |
| vimentin | AGGCAAAGCAGGAGTCCACTGA | ATCTGGCGTTCCAGGGACTCAT |
| snail | TGCCCTCAAGATGCACATCCGA | GGGACAGGAGAAGGGCTTCTC |
| twist | GCCAGGTACATCGACTTCCTCT | TCCATCCTCCAGACCGAGAAGG |
| RHOA | GCAGGTAGAGTTGGCTTTATGG | CTTGTGTGCTCATCATTCCGA |
| RHOC | AAGACGAGCACACCAGGAGAGA | TTGGCTGAGCACTCAAGGTAGC |
| ROCK1 | GAAACAGTGTTCCATGCTAGACG | GCCGCTTATTTGATTCCTGCTCC |
| β-actin | CTGGAACGGTGAAGGTGACA | AAGGGACTTCCTGTAACAACGCA |
表2 RT-qPCR引物序列
Tab 2 Sequences of primers for RT-qPCR
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| GPR87 | GGAGGCGACATCAATGCAG | AAATGAAAGTAAAGAACGATTTTGTGT |
| MMP2 | GCTGGAGACAAATTCTGGAGATACA | GTATCGAAGGCAGTGGAGAGGA |
| MMP7 | GAGTGAGCTACAGTGGGAACA | CTATGACGCGGGAGTTTAACAT |
| MMP9 | GCCACTACTGTGCCTTTGAGTC | CCCTCAGAGAATCGCCAGTACT |
| E-cadherin | GCCTCCTGAAAAGAGAGTGGAAG | TGGCAGTGTCTCTCCAAATCCG |
| N-cadherin | CCTCCAGAGTTTACTGCCATGAC | GTAGGATCTCCGCCACTGATTC |
| vimentin | AGGCAAAGCAGGAGTCCACTGA | ATCTGGCGTTCCAGGGACTCAT |
| snail | TGCCCTCAAGATGCACATCCGA | GGGACAGGAGAAGGGCTTCTC |
| twist | GCCAGGTACATCGACTTCCTCT | TCCATCCTCCAGACCGAGAAGG |
| RHOA | GCAGGTAGAGTTGGCTTTATGG | CTTGTGTGCTCATCATTCCGA |
| RHOC | AAGACGAGCACACCAGGAGAGA | TTGGCTGAGCACTCAAGGTAGC |
| ROCK1 | GAAACAGTGTTCCATGCTAGACG | GCCGCTTATTTGATTCCTGCTCC |
| β-actin | CTGGAACGGTGAAGGTGACA | AAGGGACTTCCTGTAACAACGCA |
| ID | Padj | P value | logFC | Gene symbol |
|---|---|---|---|---|
| 219936_s_at | 3.47×10-5 | 2.32×10-7 | 2.43 | GPR87 |
| 212094_at | 1.24×10-5 | 4.70×10-8 | 2.36 | PEG10 |
| 209278_s_at | 1.63×10-2 | 1.53×10-3 | 2.22 | TFPI2 |
| 227506_at | 1.25×10-5 | 5.17×10-8 | 2.14 | SLC16A9 |
| 206023_at | 3.69×10-4 | 6.62×10-6 | 2.12 | NMU |
| 205048_s_at | 3.22×10-3 | 1.44×10-4 | 2.02 | PSPH |
| 220393_at | 7.91×10-3 | 5.37×10-4 | -2.05 | LGSN |
| 213432_at | 1.33×10-3 | 3.96×10-5 | -2.17 | MUC5B |
| 225728_at | 2.93×10-4 | 4.65×10-6 | -2.23 | SORBS2 |
| 203824_at | 1.38×10-3 | 4.19×10-5 | -2.25 | TSPAN8 |
表3 与NSCLC转移及预后相关的差异表达基因
Tab 3 DEGs associated with metastasis and prognosis in NSCLC
| ID | Padj | P value | logFC | Gene symbol |
|---|---|---|---|---|
| 219936_s_at | 3.47×10-5 | 2.32×10-7 | 2.43 | GPR87 |
| 212094_at | 1.24×10-5 | 4.70×10-8 | 2.36 | PEG10 |
| 209278_s_at | 1.63×10-2 | 1.53×10-3 | 2.22 | TFPI2 |
| 227506_at | 1.25×10-5 | 5.17×10-8 | 2.14 | SLC16A9 |
| 206023_at | 3.69×10-4 | 6.62×10-6 | 2.12 | NMU |
| 205048_s_at | 3.22×10-3 | 1.44×10-4 | 2.02 | PSPH |
| 220393_at | 7.91×10-3 | 5.37×10-4 | -2.05 | LGSN |
| 213432_at | 1.33×10-3 | 3.96×10-5 | -2.17 | MUC5B |
| 225728_at | 2.93×10-4 | 4.65×10-6 | -2.23 | SORBS2 |
| 203824_at | 1.38×10-3 | 4.19×10-5 | -2.25 | TSPAN8 |
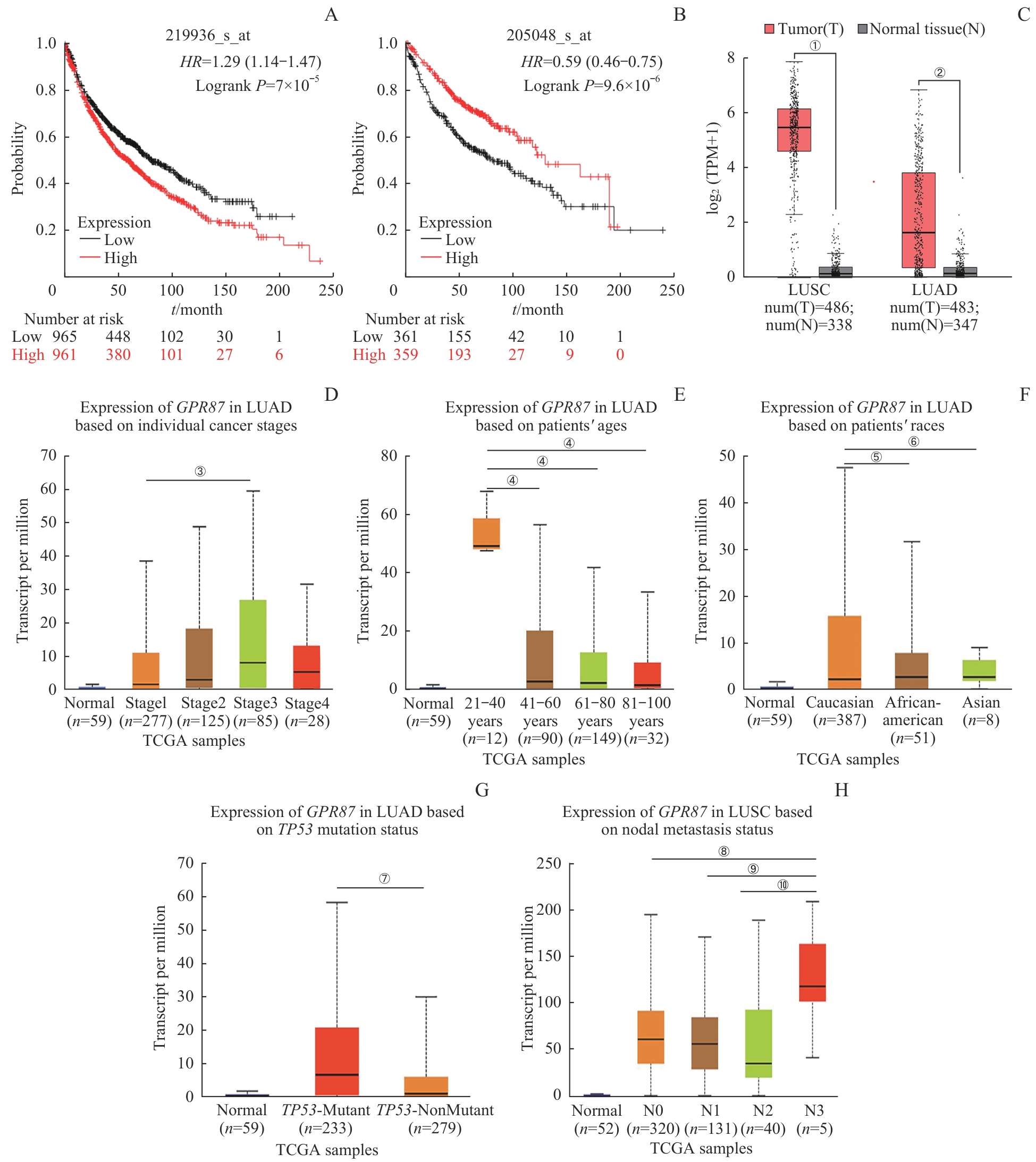
图1 GPR87 与 NSCLC的生物信息学分析Note: GPR87 (A) and PSPH (B) were found to correlate with poor survival in NSCLC population with KM plotter. GPR87 mRNA levels in LUAD and LUSC (C) were both overexpressed, according to the GEPIA database. High expression of GPR87 was associated with poor clinical stage (D), age (E), race (F) and TP53 mutation status (G) in patients with LUAD, and with lymph node metastasis in patients with LUSC (H) in UALCAN database. Error bars represent the x±s. ①P=0.032, ②P=0.026, compared with the adjacent normal tissues. ③P=0.006, compared with stage1 LUAD patients. ④P<0.001, compared with the 21?40 year-old patients. ⑤P=0.005, ⑥P<0.001, compared with Caucasian patients. ⑦P<0.001, compared with the TP53-mutant group. ⑧P=0.007, ⑨P=0.005, ⑩P=0.004, compared with the N3-lymph node metastasis patients.
Fig 1 Bioinformatics analysis of GPR87 and NSCLC
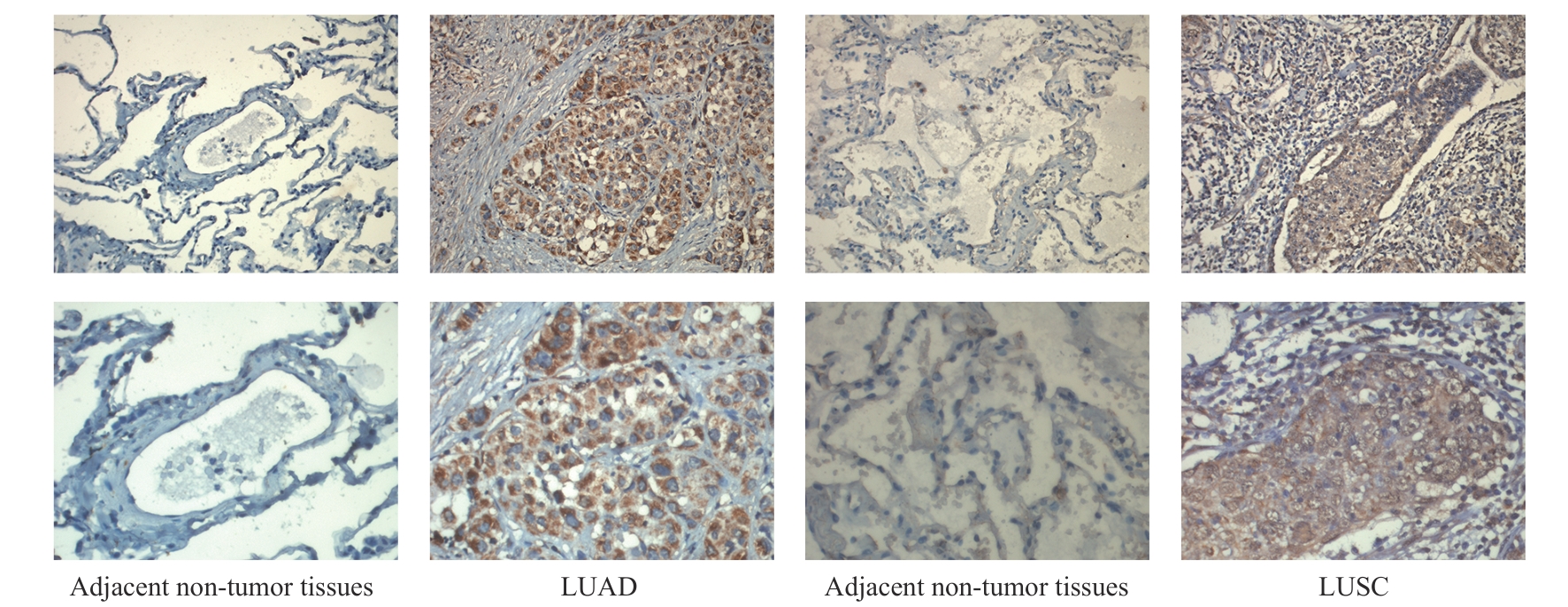
图2 GPR87蛋白在NSCLC临床样本中的表达Note: Protein expression of GPR87 in 80 NSCLC samples was upregulated compared to adjacent non-tumor tissues, as shown by IHC staining. LUAD—lung adenocarcinoma; LUSC—lung squamous cell carcinoma. Line 1, ×200; line 2, ×400.
Fig 2 Expression of GPR87 protein in NSCLC samples
| Group | Case/n | Expression of GPR87/n | χ2 | P value | |
|---|---|---|---|---|---|
| High | Low | ||||
| Tissue of NSCLC | 80 | 51 | 29 | 74.862 | <0.001 |
| Adjacent non-tumor tissues | 80 | 0 | 80 | ||
表4 GPR87在NSCLC肿瘤组织及癌旁正常组织中的蛋白表达量
Tab 4 Protein expression levels of GPR87 in NSCLC tumors and adjacent non-tumor tissues
| Group | Case/n | Expression of GPR87/n | χ2 | P value | |
|---|---|---|---|---|---|
| High | Low | ||||
| Tissue of NSCLC | 80 | 51 | 29 | 74.862 | <0.001 |
| Adjacent non-tumor tissues | 80 | 0 | 80 | ||
| Factor | Case/n | Expression of GPR87/n | χ2 | P value | |
|---|---|---|---|---|---|
| High | Low | ||||
| Subgroup | 1.524 | 0.217 | |||
| LUAD | 48 | 28 | 20 | ||
| LUSC | 32 | 23 | 9 | ||
| Age/year | 0.197 | 0.657 | |||
| ≥65 | 36 | 22 | 14 | ||
| <65 | 44 | 29 | 15 | ||
| Gender | 0.442 | 0.506 | |||
| Male | 48 | 32 | 16 | ||
| Female | 32 | 19 | 13 | ||
| Smoking | 0.548 | 0.459 | |||
| Yes/ever | 43 | 29 | 14 | ||
| No | 37 | 22 | 15 | ||
| Differentiation | 1.847 | 0.174 | |||
| Good | 11 | 5 | 6 | ||
| Middle/poor | 69 | 46 | 23 | ||
| Stage | 10.498 | 0.001 | |||
| Ⅰ/Ⅱ | 65 | 36 | 29 | ||
| Ⅲ/Ⅳ | 15 | 15 | 0 | ||
| Lymphatic metastasis | 5.485 | 0.019 | |||
| No | 50 | 27 | 23 | ||
| Yes | 30 | 24 | 6 | ||
表5 GPR87与NSCLC患者的临床相关性分析
Tab 5 The relevance of GPR87 expression to clinicopathological characteristics in NSCLC patients
| Factor | Case/n | Expression of GPR87/n | χ2 | P value | |
|---|---|---|---|---|---|
| High | Low | ||||
| Subgroup | 1.524 | 0.217 | |||
| LUAD | 48 | 28 | 20 | ||
| LUSC | 32 | 23 | 9 | ||
| Age/year | 0.197 | 0.657 | |||
| ≥65 | 36 | 22 | 14 | ||
| <65 | 44 | 29 | 15 | ||
| Gender | 0.442 | 0.506 | |||
| Male | 48 | 32 | 16 | ||
| Female | 32 | 19 | 13 | ||
| Smoking | 0.548 | 0.459 | |||
| Yes/ever | 43 | 29 | 14 | ||
| No | 37 | 22 | 15 | ||
| Differentiation | 1.847 | 0.174 | |||
| Good | 11 | 5 | 6 | ||
| Middle/poor | 69 | 46 | 23 | ||
| Stage | 10.498 | 0.001 | |||
| Ⅰ/Ⅱ | 65 | 36 | 29 | ||
| Ⅲ/Ⅳ | 15 | 15 | 0 | ||
| Lymphatic metastasis | 5.485 | 0.019 | |||
| No | 50 | 27 | 23 | ||
| Yes | 30 | 24 | 6 | ||
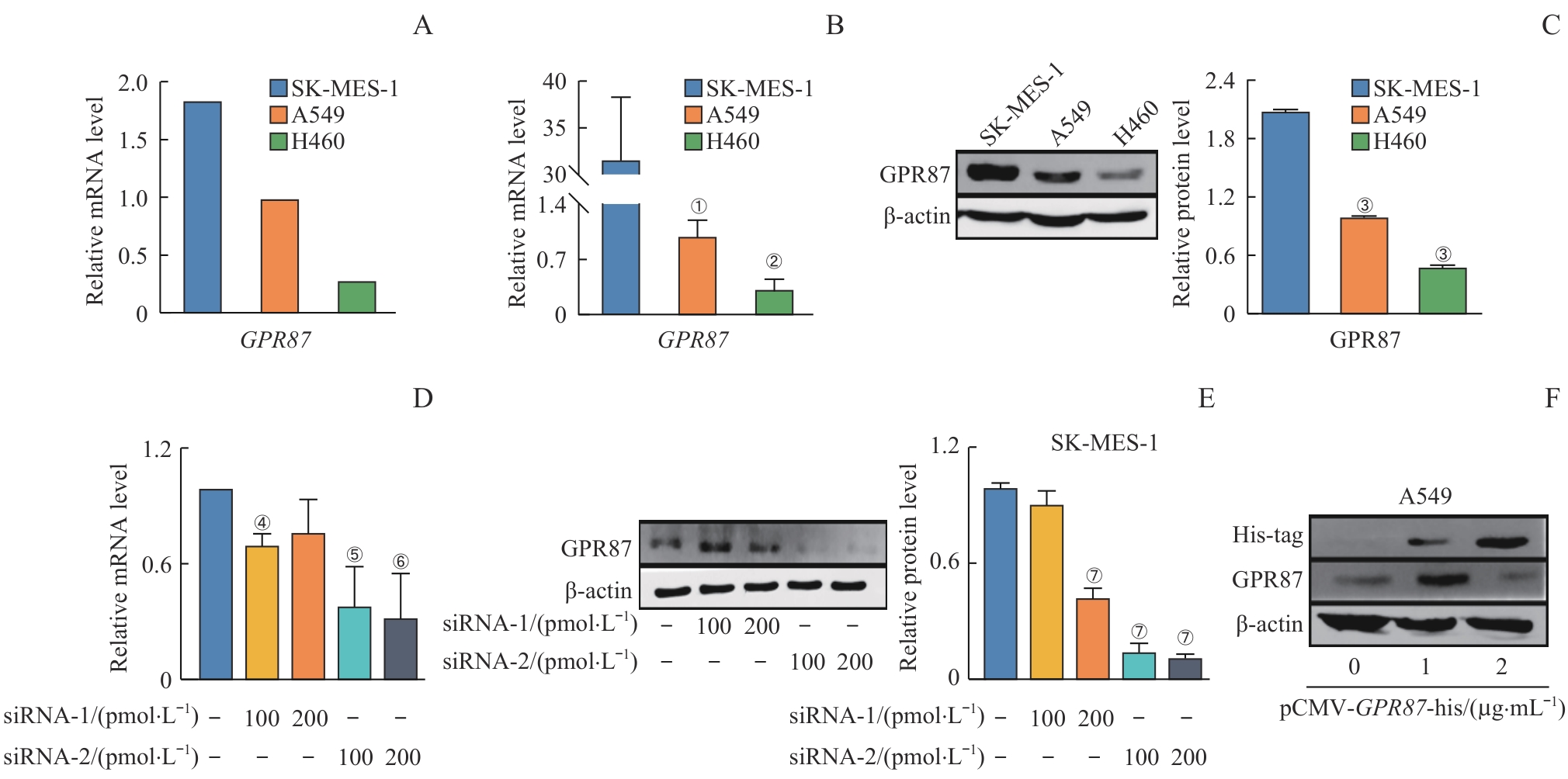
图3 GPR87 mRNA和蛋白在不同NSCLC细胞模型中的表达分析Note: Relative mRNA expression of GPR87 in SK-MES-1 and A549 cells, as predicted by CCLE (A), analyzed by RT-qPCR analysis (B), and relative protein expression of GPR87 by Western blotting analysis (C). Relative mRNA expression of GPR87 by RT-qPCR analysis (D) and relative protein expression of GPR87 by Western blotting analysis (E) in SK-MES-1 cells treated with siRNA-GPR87. GPR87 protein expression in A549 cells treated with pCMV-GPR87-his by Western blotting analysis (F). Error bars represent the x±s. ①P=0.037, ②P=0.008, compared with SK-MES-1. ③P<0.001, compared with SK-MES-1. ④P=0.008, ⑤P=0.005, ⑥P=0.006, compared with control group. ⑦P<0.001, compared with control group.
Fig 3 Analysis of GPR87 mRNA and protein expression in NSCLC cell models
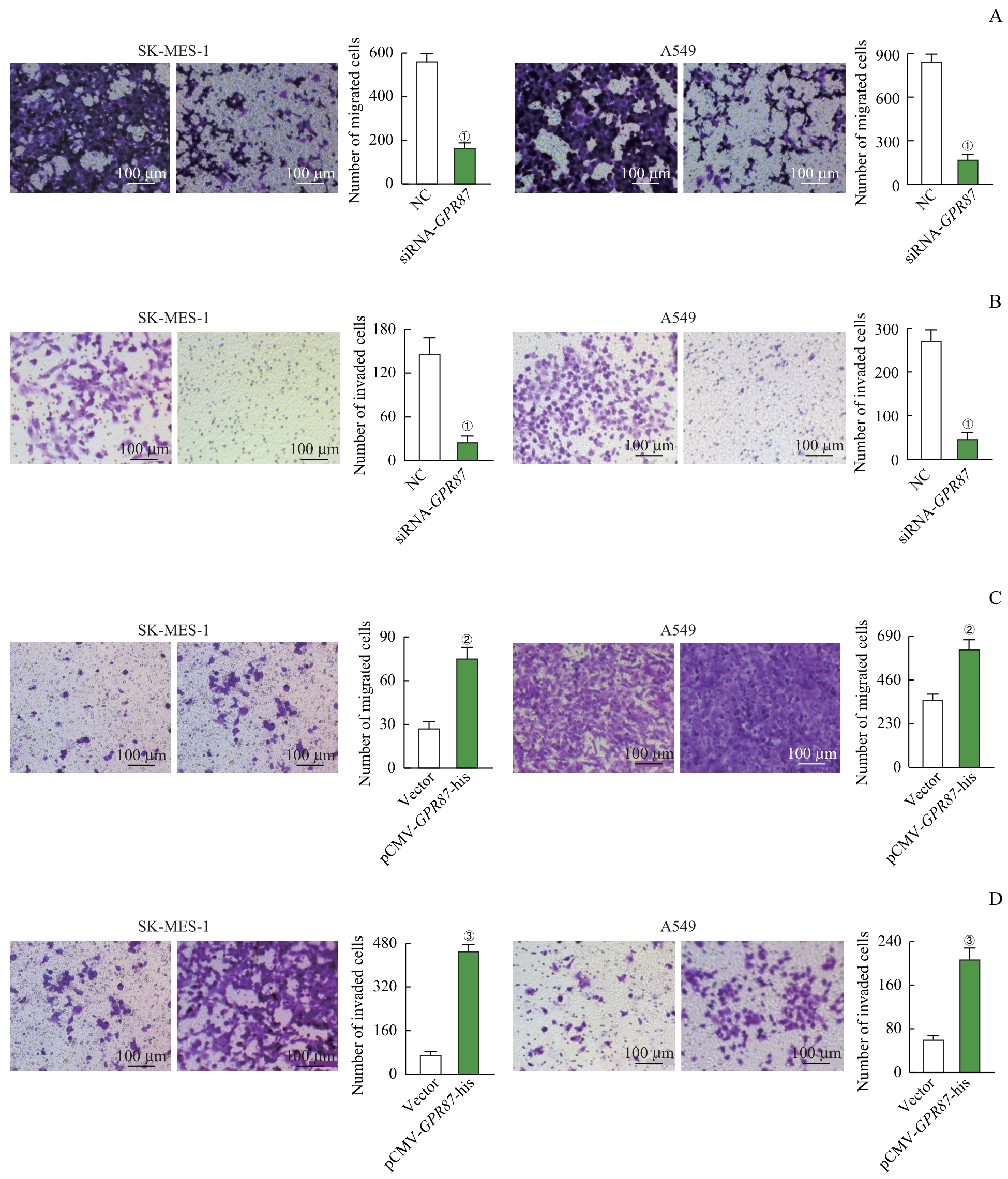
图4 Transwell检测 GPR87 对NSCLC细胞迁移和侵袭的影响Note: Representative images (×200) and quantification of migrated (A) and invaded (B) cells, analyzed by using the Transwell matrix penetration assay in SK-MES-1 and A549 cells treated with siRNA-GPR87. Representative images (×200) and quantification of migrated (C) and invaded (D) cells, analyzed by Transwell matrix penetration assay in SK-MES-1 and A549 cells treated with pCMV-GPR87-his. Error bars represent the x±s. ①P<0.001, compared with the NC group. ②P=0.008, compared with the Vector group. ③P<0.001, compared with the Vector group.
Fig 4 Effect of GPR87 on migration and invasion in NSCLC cell lines by the Transwell assay
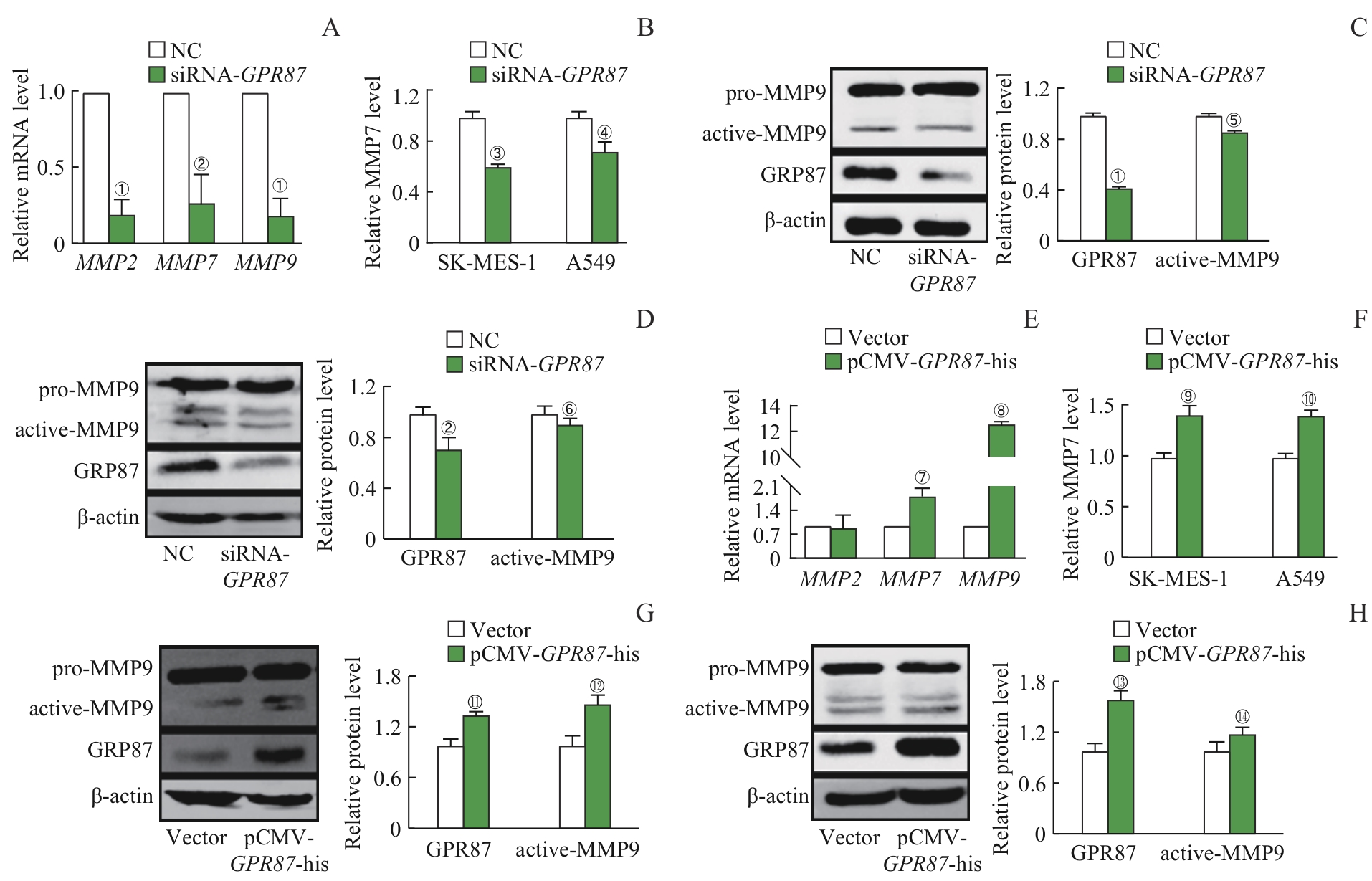
图5 GPR87 对NSCLC细胞MMPs表达的影响Note: A. The mRNA expression of MMP2, MMP7 and MMP9 in SK-MES-1 cells treated with siRNA-GPR87 was decreased, as detected by RT-qPCR. B. The protein expression of MMP7 in the supernatant of SK-MES-1 and A549 cells treated with siRNA-GPR87 was decreased, as detected by ELISA. The protein expression of active-MMP9 and GPR87 was decreased in SK-MES-1 (C) and A549 cells (D) treated with siRNA-GPR87, as detected by Western blotting. E. The mRNA expression of MMP2, MMP7 and MMP9 was increased in A549 cells treated with pCMV-GPR87-his, as detected by RT-qPCR. F. The protein expression of MMP7 in the supernatant of SK-MES-1 and A549 cells treated with pCMV-GPR87-his was increased, as detected by ELISA. The protein expression of active-MMP9 and GPR87 was increased in A549 (G) and SK-MES-1 cells (H) treated with pCMV-GPR87-his, as detected by Western blotting. Error bars represent the x±s. ①P<0.001, ②P=0.008, ③P=0.013,④P=0.041, ⑤P=0.009, ⑥P=0.043, compared with the NC group. ⑦P=0.007, ⑧P<0.001, ⑨P=0.003, ⑩P=0.002, 11P=0.005, 12P=0.008, 13P=0.001, 14P=0.021, compared with the Vector group.
Fig 5 Effect of GPR87 on the expression of MMPs in NSCLC cell lines
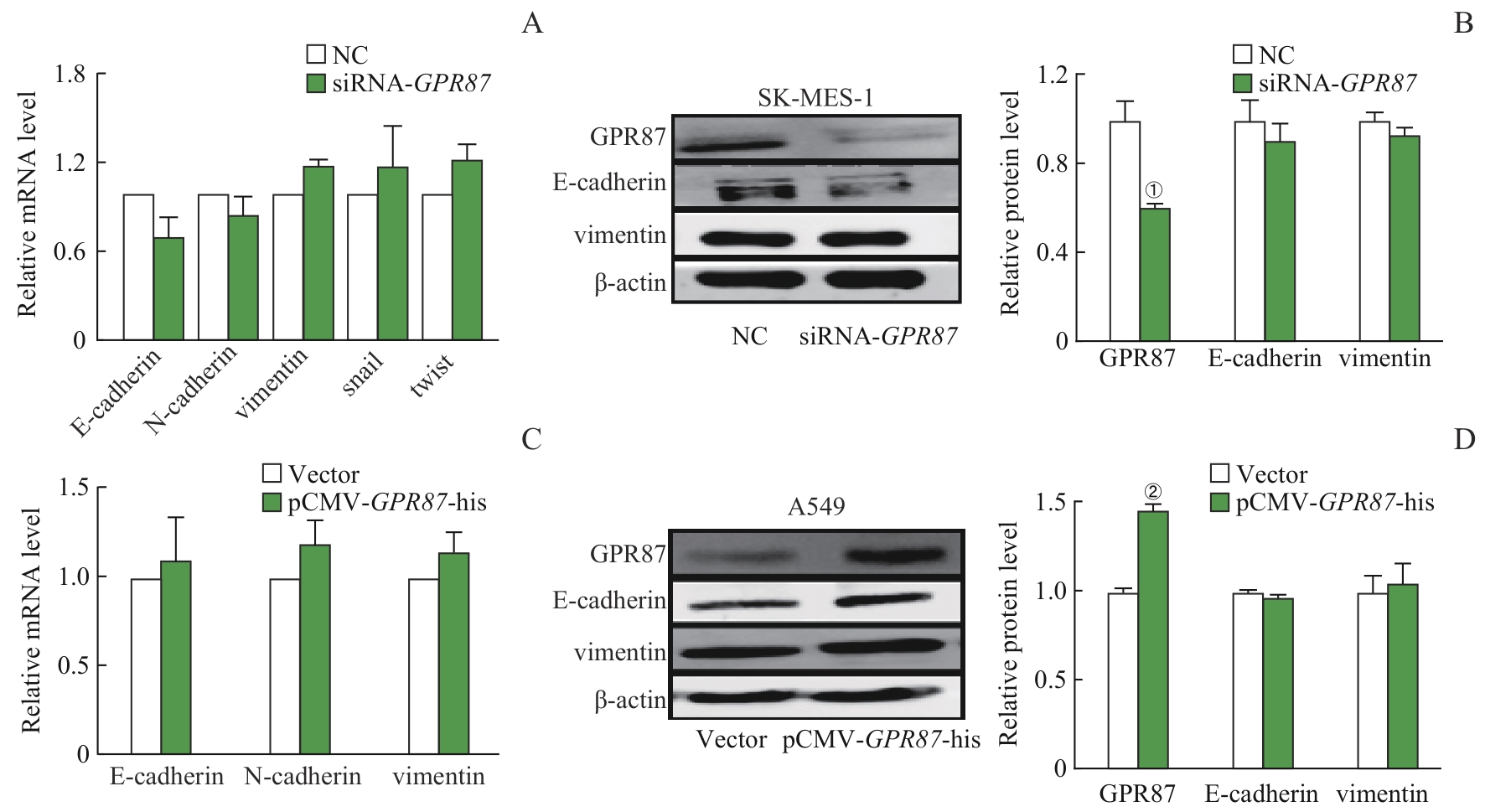
图6 GRP87 对NSCLC细胞EMT的影响Note: The mRNA expression (A) of E-cadherin, N-cadherin, vimentin, snail, and twist, and the protein expression (B) of E-cadherin and vimentin did not change in SK-MES-1 cells treated with siRNA-GPR87. The mRNA expression of E-cadherin, N-cadherin, and vimentin (C), and the protein expression of E-cadherin and vimentin (D) did not change in A549 cells treated with pCMV-GPR87-his. Error bars represent the x±s. ①P<0.001, compared with the NC group. ②P=0.008, compared with the Vector group.
Fig 6 Effect of GPR87 on EMT in NSCLC cell lines
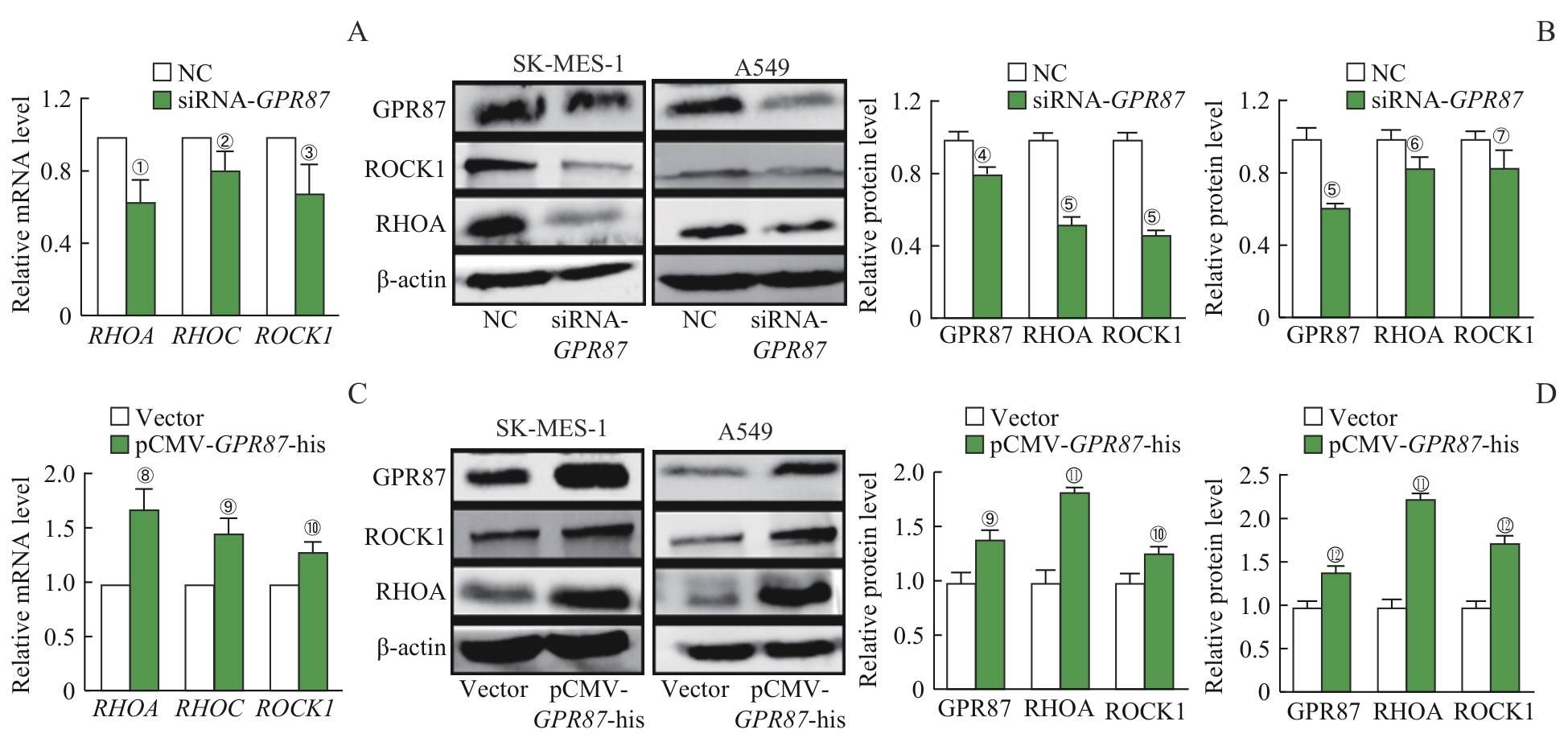
图7 GPR87 对NSCLC细胞RHO/ROCK通路的影响Note: A. The mRNA expression of RHOA, RHOC and ROCK1 was decreased in SK-MES-1 cells treated with siRNA-GPR87. B. The protein expression of RHOA and ROCK1 in SK-MES-1 and A549 cells treated with siRNA-GPR87 was decreased, as detected by Western blotting. C. The mRNA expression of RHOA, RHOC and ROCK1 in A549 cells treated with pCMV-GPR87-his was increased. D. The protein expression of RHOA and ROCK1 was increased in SK-MES-1 and A549 cells treated with pCMV-GPR87-his, as detected by Western blotting. Error bars represent the x±s. ①P=0.008, ②P=0.035, ③P=0.041, ④P=0.005, ⑤P<0.001, ⑥P=0.019, ⑦P=0.026, compared with the NC group.⑧P=0.003, ⑨P=0.002, ⑩P=0.007, 11P<0.001, 12P=0.008, compared with the Vector group.
Fig 7 Effect of GPR87 on the RHO/ROCK signaling pathway in NSCLC cell lines
| 1 | SIEGEL R L, MILLER K D, FUCHS H E, et al. Cancer statistics, 2022[J]. CA Cancer J Clin, 2022,72(1):7-33. |
| 2 | CAI Z J, ZHAN P, SONG Y, et al. Safety and efficacy of retreatment with immune checkpoint inhibitors in non-small cell lung cancer: a systematic review and meta-analysis[J]. Transl Lung Cancer Res, 2022, 11(8): 1555-1566. |
| 3 | YE Z C, HUANG Y M, KE J H, et al. Breakthrough in targeted therapy for non-small cell lung cancer[J]. Biomed Pharmacother, 2021, 133: 111079. |
| 4 | PRIESTLEY P, BABER J, LOLKEMA M P, et al. Pan-cancer whole-genome analyses of metastatic solid tumours[J]. Nature, 2019, 575(7781): 210-216. |
| 5 | BAI R, ZHANG J G, HE F J, et al. GPR87 promotes tumor cell invasion and mediates the immunogenomic landscape of lung adenocarcinoma[J]. Commun Biol, 2022, 5(1): 663. |
| 6 | KITA Y, GO T, NAKASHIMA N, et al. Inhibition of cell-surface molecular GPR87 with GPR87-suppressing adenoviral vector disturb tumor proliferation in lung cancer cells[J]. Anticancer Res, 2020, 40(2): 733-741. |
| 7 | PARK S M, CHOI E Y, BAE, et al. Histone variant H3F3A promotes lung cancer cell migration through intronic regulation[J]. Nat Commun, 2016, 7: 12914. |
| 8 | LEE S, CHO M, PARK B, et al. Finding miRNA-RNA network biomarkers for predicting metastasis and prognosis in cancer[J]. Int J Mol Sci, 2023, 24(5): 5052. |
| 9 | GUTKIND J S, KOSTENIS E. Arrestins as rheostats of GPCR signalling[J]. Nat Rev Mol Cell Biol, 2018, 19(10): 615-616. |
| 10 | CHAUDHARY P K, KIM S. An insight into GPCR and G-proteins as cancer drivers[J]. Cells, 2021, 10(12): 3288. |
| 11 | SIDDHARTHA R, GARG M. Molecular and clinical insights of matrix metalloproteinases into cancer spread and potential therapeutic interventions[J]. Toxicol Appl Pharmacol, 2021, 426: 115593. |
| 12 | MERCHANT N, NAGARAJU G P, RAJITHA B, et al. Matrix metalloproteinases: their functional role in lung cancer[J]. Carcinogenesis, 2017, 38(8): 766-780. |
| 13 | ALQURASHI Y E, AL-HETTY H R A K, RAMAIAH P, et al. Harnessing function of EMT in hepatocellular carcinoma: from biological view to nanotechnological standpoint[J]. Environ Res, 2023, 227: 115683. |
| 14 | MANSHOURI R, COYAUD E, KUNDU S T, et al. ZEB1/NuRD complex suppresses TBC1D2b to stimulate E-cadherin internalization and promote metastasis in lung cancer[J]. Nat Commun, 2019, 10(1): 5125. |
| 15 | PASTUSHENKO I, BLANPAIN C. EMT transition states during tumor progression and metastasis[J]. Trends Cell Biol, 2019, 29(3): 212-226. |
| 16 | TULCHINSKY E, DEMIDOV O, KRIAJEVSKA M, et al. EMT: a mechanism for escape from EGFR-targeted therapy in lung cancer[J]. Biochim Biophys Acta Rev Cancer, 2019, 1871(1): 29-39. |
| 17 | GUAN G Z, CANNON R D, COATES D E, et al. Effect of the rho-kinase/ROCK signaling pathway on cytoskeleton components[J]. Genes, 2023, 14(2): 272. |
| 18 | ZAKARIA M A, RAJAB N F, CHUA E W, et al. Roles of Rho-associated kinase in lung cancer (Review)[J]. Int J Oncol, 2021, 58(2): 185-198. |
| 19 | NISS ARFELT K, FARES S, SPARRE-ULRICH A H, et al. Signaling via G proteins mediates tumorigenic effects of GPR87[J]. Cell Signal, 2017, 30: 9-18. |
| 20 | JEONG K J, PARK S Y, CHO K H, et al. The Rho/ROCK pathway for lysophosphatidic acid-induced proteolytic enzyme expression and ovarian cancer cell invasion[J]. Oncogene, 2012, 31(39): 4279-4289. |
| 21 | GONG H, ZHOU L, KHELFAT L, et al. Rho-associated protein kinase (ROCK) promotes proliferation and migration of PC-3 and DU145 prostate cancer cells by targeting LIM kinase 1 (LIMK1) and matrix metalloproteinase-2 (MMP2)[J]. Med Sci Monit, 2019, 25: 3090-3099. |
| [1] | 杨娜, 刘俊丽, 白静, 杨思怡, 韩继明, 张华华. HENMT1通过激活PI3K-AKT-mTOR信号通路促进胃癌的增殖与迁移[J]. 上海交通大学学报(医学版), 2025, 45(6): 717-726. |
| [2] | 张先洲, 杜凤麟, 吴雷, 任逸喆, 赵明娜, 娄加陶. OGT通过ERK信号通路促进非小细胞肺癌增殖的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(10): 1288-1297. |
| [3] | 李想, 魏鸣, 吴文曦, 罗小琴, 姚彪, 伍思宇. 芦丁对骨肉瘤生长和转移的体内外抑制作用[J]. 上海交通大学学报(医学版), 2025, 45(1): 20-28. |
| [4] | 杨涵, 雷浩, 徐彼得, 吴淏, 马寻君, 皇艳波, 毛远青, 张经纬, 王金武. 透视立体影像分析技术在假体无菌性松动检测中的应用研究[J]. 上海交通大学学报(医学版), 2024, 44(9): 1061-1068. |
| [5] | 钱立恒, 温凯玲, 廖颖娜, 李书鑫, 聂惠贞. 分选链接蛋白1抑制结直肠癌细胞增殖和迁移的作用和机制研究[J]. 上海交通大学学报(医学版), 2024, 44(9): 1124-1135. |
| [6] | 韩依杉, 徐梓淇, 陶梦玉, 范广建, 余波. PRMT6促进乳腺癌细胞的增殖和迁移[J]. 上海交通大学学报(医学版), 2024, 44(8): 999-1010. |
| [7] | 朱鸣阳, 许元元, 任江浩, 黄嘉正, 李若楠, 谭强. 以磨玻璃结节为表现的肺腺癌亚肺叶切除研究综述[J]. 上海交通大学学报(医学版), 2024, 44(7): 922-927. |
| [8] | 谭露, 沈少明, 何平. 低氧诱导的长链非编码RNA 68在肝癌中的功能及其作用机制[J]. 上海交通大学学报(医学版), 2024, 44(6): 702-712. |
| [9] | 王桂杰, 杜传冲, 陆叶, 赵健, 沈勰, 金冬林, 耿佳财. 多发伤患者血清高迁移率族蛋白B1及可溶性髓样细胞触发受体-1水平变化及预后意义[J]. 上海交通大学学报(医学版), 2024, 44(3): 350-357. |
| [10] | 李虎虓, 李笑甜, 赵旭日, 张桓瑜, 周薇, 宋忠臣. 牙龈素提取物对口腔鳞癌细胞HN6生物学特性的影响[J]. 上海交通大学学报(医学版), 2024, 44(2): 161-168. |
| [11] | 任怡璇, 陈诚, 蔡铭慈, 陈嘉敏, 杨欣欣, 王超, 林晓珠, 程澍, 江旭峰, 陈东旭. 套细胞淋巴瘤在18F-FDG PET/CT中的显像特征及细胞形态学分型[J]. 上海交通大学学报(医学版), 2024, 44(12): 1561-1569. |
| [12] | 罗蓝鸽, 郑超, 雷鸣. 癌-睾丸抗原CT57促进肝癌细胞增殖、侵袭、迁移和上皮间质转化[J]. 上海交通大学学报(医学版), 2024, 44(11): 1335-1346. |
| [13] | 高珂星, 廖春华, 李昇泽, 马双羽, 黄雷. 黏蛋白1调控肿瘤细胞恶性特征的功能位点分析[J]. 上海交通大学学报(医学版), 2024, 44(11): 1370-1382. |
| [14] | 陈宁黛, 周冰倩, 陈哲逸, 陈诗宇, 郑英霞. 赖氨酸去甲基化酶5C对肾癌转移的作用研究[J]. 上海交通大学学报(医学版), 2023, 43(5): 571-579. |
| [15] | 黄华艳, 徐张闻笛, 夏立亮, 虞永峰, 陆舜. 表皮生长因子受体突变型晚期非小细胞肺癌免疫治疗的研究进展[J]. 上海交通大学学报(医学版), 2023, 43(5): 611-618. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||