
上海交通大学学报(医学版) ›› 2024, Vol. 44 ›› Issue (11): 1460-1465.doi: 10.3969/j.issn.1674-8115.2024.11.014
• 综述 • 上一篇
收稿日期:2024-03-31
接受日期:2024-07-16
出版日期:2024-11-28
发布日期:2024-11-28
通讯作者:
马皎
E-mail:borisgao0219@sjtu.edu.cn;drjiaoma@shsmu.edu.cn
作者简介:高翌轩(2004—),男,本科生;电子信箱:borisgao0219@sjtu.edu.cn。
基金资助:
GAO Yixuan1( ), ZHANG Yichi1, DAI Luyan1, MA Jiao2(
), ZHANG Yichi1, DAI Luyan1, MA Jiao2( )
)
Received:2024-03-31
Accepted:2024-07-16
Online:2024-11-28
Published:2024-11-28
Contact:
MA Jiao
E-mail:borisgao0219@sjtu.edu.cn;drjiaoma@shsmu.edu.cn
Supported by:摘要:
黏蛋白型O-糖基化是蛋白质中最常见的翻译后修饰之一,可以改变蛋白质的空间构象与生物学功能,在细胞信号转导、细胞黏附、免疫应答等生物学过程中发挥着关键作用。而多肽N-乙酰半乳糖胺转移酶3(polypeptide N-acetylgalactosaminyltransferase 3,GALNT3)作为黏蛋白型O-糖基化的起始酶,在维持人类细胞和组织的稳态中具有重要意义,而其功能失调已被发现在钙磷代谢紊乱、动脉粥样硬化等多种疾病中发挥作用。此外,GALNT3被发现在结直肠癌、肺癌、卵巢癌等多种肿瘤组织中异常表达,且与患者的临床病理特征和较差预后相关,可以作为肿瘤早期诊断和预后评估的潜在标志物。进一步研究表明,GALNT3既可通过调控糖基化水平,降低肿瘤细胞之间的黏附水平,也可通过激活多条代谢相关通路,促进肿瘤细胞的侵袭转移。该文就GALNT3在恶性肿瘤发生发展中的作用进行综述,并分析了靶向GALNT3开发抗肿瘤药物的前景与挑战。
中图分类号:
高翌轩, 张亦弛, 戴鹭俨, 马皎. GALNT3作为潜在肿瘤分子标志物及药物靶点的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(11): 1460-1465.
GAO Yixuan, ZHANG Yichi, DAI Luyan, MA Jiao. Research progress of GALNT3 as a potential tumor molecular marker and drug target[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(11): 1460-1465.
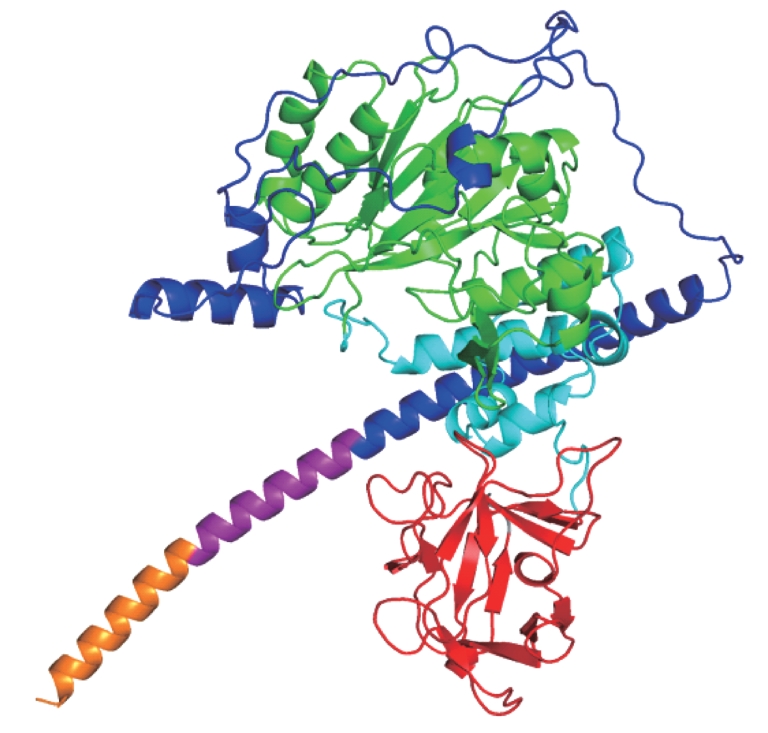
图1 GALNT3蛋白的结构示意图Note: The orange part represents the N-terminal cytoplasm tail, the purple part represents the transmembrane domain, the blue part is the stem region, the green part is the catalytic domain, the cyan part is the linker region, the red part represents the lectin domain, and the grey part is the C-terminal residue. The figure was prepared using PyMOL software.
Fig 1 Schematic diagram of structural conformation of GALNT3 protein
| 1 | VAN DEN STEEN P, RUDD P M, DWEK R A, et al. Concepts and principles of O-linked glycosylation[J]. Crit Rev Biochem Mol Biol, 1998, 33(3): 151-208. |
| 2 | ESMAIL S, MANOLSON M F. Advances in understanding N-glycosylation structure, function, and regulation in health and disease[J]. Eur J Cell Biol, 2021, 100(7/8): 151186. |
| 3 | WANDALL H H, NIELSEN M A I, KING-SMITH S, et al. Global functions of O-glycosylation: promises and challenges in O-glycobiology[J]. FEBS J, 2021, 288(24): 7183-7212. |
| 4 | GARAY Y C, CEJAS R B, LORENZ V, et al. Polypeptide N-acetylgalactosamine transferase 3: a post-translational writer on human health[J]. J Mol Med, 2022, 100(10): 1387-1403. |
| 5 | ČAVAL T, ALISSON-SILVA F, SCHWARZ F. Roles of glycosylation at the cancer cell surface: opportunities for large scale glycoproteomics[J]. Theranostics, 2023, 13(8): 2605-2615. |
| 6 | MAGALHÃES A, DUARTE H O, REIS C A. The role of O-glycosylation in human disease[J]. Mol Aspects Med, 2021, 79: 100964. |
| 7 | LI S D, QI Y, HUANG Y R, et al. Exosome-derived SNHG16 sponging miR-4500 activates HUVEC angiogenesis by targeting GALNT1 via PI3K/Akt/mTOR pathway in hepatocellular carcinoma[J]. J Physiol Biochem, 2021, 77(4): 667-682. |
| 8 | LIU S Y, SHUN C T, HUNG K Y, et al. Mucin glycosylating enzyme GALNT2 suppresses malignancy in gastric adenocarcinoma by reducing MET phosphorylation[J]. Oncotarget, 2016, 7(10): 11251-11262. |
| 9 | LIU C, LI Z, XU L, et al. GALNT6 promotes breast cancer metastasis by increasing mucin-type O-glycosylation of α2M[J]. Aging, 2020, 12(12): 11794-11811. |
| 10 | PENG X D, CHEN X R, ZHU X T, et al. GALNT6 knockdown inhibits the proliferation and migration of colorectal cancer cells and increases the sensitivity of cancer cells to 5-FU[J]. J Cancer, 2021, 12(24): 7413-7421. |
| 11 | LI H W, LIU M B, JIANG X, et al. GALNT14 regulates ferroptosis and apoptosis of ovarian cancer through the EGFR/mTOR pathway[J]. Future Oncol, 2022, 18(2): 149-161. |
| 12 | MARIMUTHU S, BATRA S K, PONNUSAMY M P. Pan-cancer analysis of altered glycosyltransferases confers poor clinical outcomes[J]. Clin Transl Discov, 2022, 2(2): e100. |
| 13 | MAO C Z, ZHUANG S M, XIA Z J, et al. Pan-cancer analysis of GALNTs expression identifies a prognostic of GALNTs feature in low grade glioma[J]. J Leukoc Biol, 2022, 112(4): 887-899. |
| 14 | RODRIGUEZ E, BOELAARS K, BROWN K, et al. Analysis of the glyco-code in pancreatic ductal adenocarcinoma identifies glycan-mediated immune regulatory circuits[J]. Commun Biol, 2022, 5(1): 41. |
| 15 | MEIJLINK F, CURRAN T, MILLER A D, et al. Removal of a 67-base-pair sequence in the noncoding region of protooncogene fos converts it to a transforming gene[J]. Proc Natl Acad Sci U S A, 1985, 82(15): 4987-4991. |
| 16 | HO B B, BERGWITZ C. FGF23 signalling and physiology[J]. J Mol Endocrinol, 2021, 66(2): R23-R32. |
| 17 | HASSAN N, GREGSON C L, TANG H T, et al. Rare and common variants in GALNT3 may affect bone mass independently of phosphate metabolism[J]. J Bone Miner Res, 2023, 38(5): 678-691. |
| 18 | ITO N, FUKUMOTO S. Congenital hyperphosphatemic conditions caused by the deficient activity of FGF23[J]. Calcif Tissue Int, 2021, 108(1): 104-115. |
| 19 | GUO L W, LI D, LI M T, et al. Variant in GALNT3 gene linked with reduced coronary artery disease risk in Chinese population[J]. DNA Cell Biol, 2017, 36(7): 529-534. |
| 20 | WANG Y K, LI S J, ZHOU L L, et al. GALNT3 protects against vascular calcification by reducing oxidative stress and apoptosis of smooth muscle cells[J]. Eur J Pharmacol, 2023, 939: 175447. |
| 21 | GUO L W, WANG L Y, LI H F, et al. Down regulation of GALNT3 contributes to endothelial cell injury via activation of p38 MAPK signaling pathway[J]. Atherosclerosis, 2016, 245: 94-100. |
| 22 | BENNETT E P, MANDEL U, CLAUSEN H, et al. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family[J]. Glycobiology, 2012, 22(6): 736-756. |
| 23 | RAGHU D, MOBLEY R J, SHENDY N A M, et al. GALNT3 maintains the epithelial state in trophoblast stem cells[J]. Cell Rep, 2019, 26(13): 3684-3697.e7. |
| 24 | PELUSO G, TIAN E, ABUSLEME L, et al. Loss of the disease-associated glycosyltransferase Galnt3 alters Muc10 glycosylation and the composition of the oral microbiome[J]. J Biol Chem, 2020, 295(5): 1411-1425. |
| 25 | NYGAARD M B, HERLIHY A S, JEANNEAU C, et al. Expression of the O-glycosylation enzyme GalNAc-T3 in the equatorial segment correlates with the quality of spermatozoa[J]. Int J Mol Sci, 2018, 19(10): 2949. |
| 26 | BAGDONAITE I, PALLESEN E M, YE Z L, et al. O-glycan initiation directs distinct biological pathways and controls epithelial differentiation[J]. EMBO Rep, 2020, 21(6): e48885. |
| 27 | ONITSUKA K, SHIBAO K, NAKAYAMA Y, et al. Prognostic significance of UDP-N-acetyl-α-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-3 (GalNAc-T3) expression in patients with gastric carcinoma[J]. Cancer Sci, 2003, 94(1): 32-36. |
| 28 | KITADA S, YAMADA S, KUMA A, et al. Polypeptide N-acetylgalactosaminyl transferase 3 independently predicts high-grade tumours and poor prognosis in patients with renal cell carcinomas[J]. Br J Cancer, 2013, 109(2): 472-481. |
| 29 | WANG Z Q, BACHVAROVA M, MORIN C, et al. Role of the polypeptide N-acetylgalactosaminyltransferase 3 in ovarian cancer progression: possible implications in abnormal mucin O-glycosylation[J]. Oncotarget, 2014, 5(2): 544-560. |
| 30 | LUO D, FANG M Y, SHAO L, et al. The EMT-related genes GALNT3 and OAS1 are associated with immune cell infiltration and poor prognosis in lung adenocarcinoma[J]. Front Biosci, 2023, 28(10): 271. |
| 31 | SHIBAO K, IZUMI H, NAKAYAMA Y, et al. Expression of UDP-N-acetyl-α-D-galactosamine-polypeptide galNAc N-acetylgalactosaminyl transferase-3 in relation to differentiation and prognosis in patients with colorectal carcinoma[J]. Cancer, 2002, 94(7): 1939-1946. |
| 32 | MOCHIZUKI Y, ITO K, IZUMI H, et al. Expression of polypeptide N-acetylgalactosaminyl transferase-3 and its association with clinicopathological factors in thyroid carcinomas[J]. Thyroid, 2013, 23(12): 1553-1560. |
| 33 | BARKEER S, CHUGH S, KARMAKAR S, et al. Novel role of O-glycosyltransferases GALNT3 and B3GNT3 in the self-renewal of pancreatic cancer stem cells[J]. BMC Cancer, 2018, 18(1): 1157. |
| 34 | LIU B, PAN S M, XIAO Y, et al. LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway[J]. J Exp Clin Cancer Res, 2018, 37(1): 316. |
| 35 | SUN L X, SUN W, SONG H B, et al. MiR-885-5p inhibits proliferation and metastasis by targeting IGF2BP1 and GALNT3 in human intrahepatic cholangiocarcinoma[J]. Mol Carcinog, 2020, 59(12): 1371-1381. |
| 36 | ISHIKAWA M, KITAYAMA J, NARIKO H, et al. The expression pattern of UDP-N-acetyl-α-D-galactosamine: polypeptide N-acetylgalactosaminyl transferase-3 in early gastric carcinoma[J]. J Surg Oncol, 2004, 86(1): 28-33. |
| 37 | PARK M S, YANG A Y, LEE J E, et al. GALNT3 suppresses lung cancer by inhibiting myeloid-derived suppressor cell infiltration and angiogenesis in a TNFR and c-MET pathway-dependent manner[J]. Cancer Lett, 2021, 521: 294-307. |
| 38 | PANG X C, LI H J, GUAN F, et al. Multiple roles of glycans in hematological malignancies[J]. Front Oncol, 2018, 8: 364. |
| 39 | OLIVEIRA T, ZHANG M F, JOO E J, et al. Glycoproteome remodeling in MLL-rearranged B-cell precursor acute lymphoblastic leukemia[J]. Theranostics, 2021, 11(19): 9519-9537. |
| 40 | SUPRUNIUK K, RADZIEJEWSKA I. MUC1 is an oncoprotein with a significant role in apoptosis (Review)[J]. Int J Oncol, 2021, 59(3): 68. |
| 41 | ALOBAIDI N K, ALWAN A F, AL-REKABI A N. Assessment of LAG3 and GALNT11 gene expression in patients with chronic lymphocytic leukemia and their impact on disease progression[J]. Biochem Cell Arch, 2021, 21(1): 809-818. |
| 42 | PATSOS G, HEBBE-VITON V, ROBBE-MASSELOT C, et al. O-glycan inhibitors generate aryl-glycans, induce apoptosis and lead to growth inhibition in colorectal cancer cell lines[J]. Glycobiology, 2009, 19(4): 382-398. |
| 43 | SONG L N, LINSTEDT A D. Inhibitor of ppGalNAc-T3-mediated O-glycosylation blocks cancer cell invasiveness and lowers FGF23 levels[J]. eLife, 2017, 6: e24051. |
| 44 | KIM Y K. RNA therapy: current status and future potential[J]. Chonnam Med J, 2020, 56(2): 87-93. |
| 45 | CHENG D, CHU F F, LIANG F, et al. Downregulation of circ-RAPGEF5 inhibits colorectal cancer progression by reducing the expression of polypeptide N-acetylgalactosaminyltransferase 3 (GALNT3)[J]. Environ Toxicol, 2024, 39(8): 4249-4260. |
| 46 | ZHANG X Y, XIE K, ZHOU H H, et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance[J]. Mol Cancer, 2020, 19(1): 47. |
| 47 | TAHERIAZAM A, BAYANZADEH S D, HEYDARI FARAHANI M, et al. Non-coding RNA-based therapeutics in cancer therapy: an emphasis on Wnt/β-catenin control[J]. Eur J Pharmacol, 2023, 951: 175781. |
| 48 | ROBINSON E L, PORT J D. Utilization and potential of RNA-based therapies in cardiovascular disease[J]. JACC Basic Transl Sci, 2022, 7(9): 956-969. |
| 49 | PAUNOVSKA K, LOUGHREY D, DAHLMAN J E. Drug delivery systems for RNA therapeutics[J]. Nat Rev Genet, 2022, 23(5): 265-280. |
| 50 | PARK H, OTTE A, PARK K. Evolution of drug delivery systems: from 1950 to 2020 and beyond[J]. J Control Release, 2022, 342: 53-65. |
| 51 | KHAN M I, HOSSAIN M I, HOSSAIN M K, et al. Recent progress in nanostructured smart drug delivery systems for cancer therapy: a review[J]. ACS Appl Bio Mater, 2022, 5(3): 971-1012. |
| 52 | NARIMATSU Y, JOSHI H J, YANG Z, et al. A validated gRNA library for CRISPR/Cas9 targeting of the human glycosyltransferase genome[J]. Glycobiology, 2018, 28(5): 295-305. |
| [1] | 王梦菲, 杨守志, 乔永霞, 黄琳. 基于临床检验指标建立肺腺癌患者浸润程度判别模型[J]. 上海交通大学学报(医学版), 2024, 44(1): 98-107. |
| [2] | 朱屿倩,吴凌云. m6A甲基化修饰在血液系统恶性肿瘤中的作用研究进展[J]. 上海交通大学学报(医学版), 2020, 40(3): 396-. |
| [3] | 刘锦燕1,赵珺涛2,王恒如2,黄嘉仪2,王钰婷2,项明洁1, 2. 细胞骨架相关蛋白4对肿瘤诊断和预后的作用[J]. 上海交通大学学报(医学版), 2020, 40(2): 255-. |
| [4] | 刘利杰 1,洪西 1,黄先玉 3,罗静 3,俞建军 1, 2. 前列腺癌中巴豆酰化修饰的测定及与肿瘤分期和分级的相关性[J]. 上海交通大学学报(医学版), 2018, 38(7): 788-. |
| [5] | 张海晨 1, 2,马进 1. 基于 IMB 模型的肺部疾病肿瘤标志物检测行为调查[J]. 上海交通大学学报(医学版), 2018, 38(2): 206-. |
| [6] | 石晓东,戴钰俊,王月英. DNMT3A 在血液肿瘤中的作用#br#[J]. 上海交通大学学报(医学版), 2017, 37(9): 1276-. |
| [7] | 郑田玉,杜君,陈宁,倪培华,薛惠平 . 外泌体及其在肿瘤诊疗中的作用[J]. 上海交通大学学报(医学版), 2017, 37(7): 1046-. |
| [8] | 夏莉,王月英 . 嵌合抗原受体 T 细胞疗法及其在血液肿瘤免疫治疗中的应用[J]. 上海交通大学学报(医学版), 2017, 37(6): 822-. |
| [9] | 徐媛媛,赵洪,张宇,孙皎,顾芹,汪海东,朱君秋. 老年2型糖尿病患者5种血清肿瘤标志物的表达及影响因素分析[J]. 上海交通大学学报(医学版), 2016, 36(7): 1044-. |
| [10] | 顾文莉,吴静静. 循环miR-4677 在口腔鳞状细胞癌患者中的表达[J]. 上海交通大学学报(医学版), 2015, 35(3): 348-. |
| [11] | 蒋奕玫,赵任. 肿瘤标志物在结直肠癌诊断中的作用[J]. 上海交通大学学报(医学版), 2015, 35(11): 1734-. |
| [12] | 华子辰,朱正伦,朱正纲. 血清肿瘤标志物在胃癌诊疗中的应用[J]. 上海交通大学学报(医学版), 2014, 34(9): 1411-. |
| [13] | 王鸣明,邹丽芳,窦红菊,等. PBL结合临床路径教学模式在血液肿瘤实习教学中的应用[J]. 上海交通大学学报(医学版), 2014, 34(11): 1678-. |
| [14] | 庞淯阳, 王 婷, 陈芳源. 代谢组学技术在肿瘤标志物筛选中的应用[J]. , 2012, 32(6): 805-. |
| [15] | 张军峰, 花 荣. 微小RNA在胰腺导管腺癌诊断、治疗和预后评估中的研究进展[J]. , 2012, 32(12): 1646-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||