
上海交通大学学报(医学版) ›› 2025, Vol. 45 ›› Issue (12): 1606-1619.doi: 10.3969/j.issn.1674-8115.2025.12.006
• 论著 · 循证医学 • 上一篇
马会华1,2, 闫奎坡1( ), 刘刚1, 徐亚洲1, 张磊1, 孙彦琴1
), 刘刚1, 徐亚洲1, 张磊1, 孙彦琴1
收稿日期:2025-04-17
接受日期:2025-06-17
出版日期:2025-12-28
发布日期:2025-12-28
通讯作者:
闫奎坡,主任医师,博士;电子信箱:ykp19821122@163.com。基金资助:
MA Huihua1,2, YAN Kuipo1( ), LIU Gang1, XU Yazhou1, ZHANG Lei1, SUN Yanqin1
), LIU Gang1, XU Yazhou1, ZHANG Lei1, SUN Yanqin1
Received:2025-04-17
Accepted:2025-06-17
Online:2025-12-28
Published:2025-12-28
Contact:
YAN Kuipo, E-mail: ykp19821122@163.com.Supported by:摘要:
目的·通过孟德尔随机化(Mendelian randomization,MR)方法探讨肠道微生物群与心血管疾病(cardiovascular disease,CVD)之间的因果关系。方法·使用MiBioGen联盟提供的肠道微生物群数据(n=18 340)和IEU Open GWAS数据库提供的4种CVD(心房颤动1 030 836例、冠状动脉疾病547 261例、高血压20 526例、心力衰竭977 323例)相关的遗传位点作为工具变量。研究采用逆方差加权法(inverse variance weighted,IVW)作为主要研究方法。同时,使用Cochran's Q检验评估遗传工具变量的异质性,MR-Egger截距检验评估水平多效性,留一法评估作为工具变量的单核苷酸多态性(single-nucleotide polymorphism,SNP)对暴露和结局因果关系影响的敏感性。采用MR Steiger检验验证肠道微生物群与CVD之间的因果方向。结果·IVW法的研究结果表明:Victivallales(OR=0.939)、霍氏菌属(OR=0.939)、厌氧链球菌属(OR=0.922)、双歧杆菌科(OR=0.916)、黏胶球形菌纲(OR=0.936)、臭气杆菌属(OR=0.909)、Intestinibacter(OR=0.933)、黏胶球形菌门(OR=0.926)和双歧杆菌目(OR=0.916)对心房颤动表现为保护因素,而链状杆菌属(OR=1.057)、毛螺菌科UCG008(OR=1.051)、链球菌属(OR=1.089)和Victivallis(OR=1.038)则为危险因素;乳杆菌目(OR=0.919)和副拟杆菌属(OR=0.866)是冠状动脉疾病的保护因素,而韦荣球菌科(OR=1.065)、Lachnoclostridium(OR=1.093)、毛螺菌科(OR=1.094)、草酸杆菌属(OR=1.062)、臭气杆菌属(OR=1.160)是危险因素;Mollicutes RF9(OR=0.851),红椿菌纲(OR=0.803)、目(OR=0.803)、科(OR=0.803),以及Intestinibacter(OR=0.819)是高血压的保护因素,而Christensenellaceae R7 group(OR=1.218)、脱硫弧菌属(OR=1.167)和消化球菌科(OR=1.230)是危险因素;芽孢杆菌目(OR=0.955)和厌氧链球菌属(OR=0.899)是心力衰竭的保护因素,而瘤胃球菌UCG009(OR=1.107)、Eubacterium oxidoreducens group(OR=1.117)、月形单胞菌目(OR=1.106)、阴性杆菌目(OR=1.107)、Eubacterium eligens group(OR=1.139)和解黄酮菌属(OR=1.144)是危险因素。Cochran's Q检验显示,与CVD存在因果关系的肠道微生物群的SNP之间不存在异质性(均P>0.05);基因多效性检验未发现多效性(均P>0.05);留一法敏感性分析证实研究结果的稳健性。MR Steiger方向性检验结果支持肠道微生物群作为暴露、CVD作为结局的因果方向。结论·部分肠道微生物群对CVD存在显著的因果效应;改变其丰度可能影响CVD风险,这为基于微生物群的干预策略提供了潜在靶点。
中图分类号:
马会华, 闫奎坡, 刘刚, 徐亚洲, 张磊, 孙彦琴. 肠道微生物群与心血管疾病的因果关系评价:双向孟德尔随机化分析[J]. 上海交通大学学报(医学版), 2025, 45(12): 1606-1619.
MA Huihua, YAN Kuipo, LIU Gang, XU Yazhou, ZHANG Lei, SUN Yanqin. Causal relationship between gut microbiota and cardiovascular diseases: a bidirectional Mendelian randomization analysis[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2025, 45(12): 1606-1619.
| GWAS data ID | Disease | Sample size/n | Case/n | Control/n | Population |
|---|---|---|---|---|---|
| ebi-a-GCST006414 | AF | 1 030 836 | 60 620 | 970 216 | European |
| ebi-a-GCST005195 | CAD | 547 261 | 122 733 | 424 528 | European |
| ebi-a-GCST008036 | Hypertension | 20 526 | 11 863 | 8 663 | European |
| ebi-a-GCST009541 | HF | 977 323 | 47 309 | 930 014 | European |
表1 CVD的GWAS数据集
Tab 1 GWAS datasets for CVD
| GWAS data ID | Disease | Sample size/n | Case/n | Control/n | Population |
|---|---|---|---|---|---|
| ebi-a-GCST006414 | AF | 1 030 836 | 60 620 | 970 216 | European |
| ebi-a-GCST005195 | CAD | 547 261 | 122 733 | 424 528 | European |
| ebi-a-GCST008036 | Hypertension | 20 526 | 11 863 | 8 663 | European |
| ebi-a-GCST009541 | HF | 977 323 | 47 309 | 930 014 | European |
| Outcome | Exposure | MR method | SNP/n | β | se | OR (95%CI) | P value | Correct causal direction | P value (Steiger test) |
|---|---|---|---|---|---|---|---|---|---|
| AF | |||||||||
| Catenibacterium | IVW | 5 | 0.05 | 0.025 | 1.057 (1.005‒1.112) | 0.030 | True | 4.85×10-6 | |
| Victivallales | IVW | 8 | -0.06 | 0.023 | 0.939 (0.896‒0.984) | 0.008 | True | 1.52×10-6 | |
| Howardella | IVW | 10 | -0.06 | 0.019 | 0.939 (0.904‒0.977) | 0.001 | True | 3.04×10-5 | |
| Lachnospiraceae UCG008 | IVW | 11 | 0.04 | 0.025 | 1.051 (1.001‒1.104) | 0.047 | True | 2.52×10-7 | |
| Anaerostipes | IVW | 13 | -0.08 | 0.037 | 0.922 (0.857‒0.993) | 0.030 | True | 5.20×10-6 | |
| Bifidobacteriaceae | IVW | 11 | -0.08 | 0.034 | 0.916 (0.855‒0.980) | 0.011 | True | 1.42×10-5 | |
| Lentisphaeria | IVW | 8 | -0.06 | 0.024 | 0.936 (0.887‒0.983) | 0.003 | True | 3.96×10-6 | |
| Streptococcus | IVW | 15 | 0.08 | 0.039 | 1.089 (1.008‒1.177) | 0.029 | True | 9.37×10-6 | |
| Victivallis | IVW | 10 | 0.03 | 0.018 | 1.038 (1.001‒1.077) | 0.043 | True | 4.62×10-6 | |
| Odoribacter | IVW | 7 | -0.09 | 0.046 | 0.909 (0.831‒0.996) | 0.040 | True | 1.81×10-5 | |
| Intestinibacter | IVW | 15 | -0.06 | 0.031 | 0.933 (0.877‒0.993) | 0.028 | True | 2.28×10-6 | |
| Lentisphaerae | IVW | 9 | -0.07 | 0.022 | 0.926 (0.886‒0.968) | <0.001 | True | 5.06×10-6 | |
| Bifidobacteriales | IVW | 11 | -0.08 | 0.034 | 0.916 (0.855‒0.980) | 0.011 | True | 1.35×10-5 | |
| CAD | |||||||||
| Lactobacillales | IVW | 15 | -0.08 | 0.032 | 0.919 (0.862‒0.980) | 0.010 | True | 9.28×10-7 | |
| Veillonellaceae | IVW | 19 | 0.06 | 0.026 | 1.065 (1.011‒1.122) | 0.018 | True | 1.21×10-5 | |
| Parabacteroides | IVW | 6 | -0.14 | 0.050 | 0.866 (0.784‒0.956) | 0.004 | True | 6.31×10-5 | |
| Lachnospiraceae | IVW | 17 | 0.09 | 0.038 | 1.094 (1.012‒1.183) | 0.023 | True | 7.23×10-6 | |
| Lachnoclostridium | IVW | 13 | 0.08 | 0.039 | 1.093 (1.011‒1.181) | 0.025 | True | 9.14×10-5 | |
| Oxalobacter | IVW | 11 | 0.05 | 0.020 | 1.062 (1.019‒1.106) | 0.004 | True | 6.05×10-6 | |
| Odoribacter | IVW | 7 | 0.14 | 0.047 | 1.160 (1.056‒1.275) | 0.001 | True | 6.42×10-7 | |
| Hypertension | |||||||||
| Mollicutes RF9 | IVW | 13 | -0.16 | 0.068 | 0.851 (0.743‒0.974) | 0.019 | True | 7.23×10-5 | |
| Peptococcaceae | IVW | 9 | 0.20 | 0.086 | 1.230 (1.038‒1.458) | 0.016 | True | 2.07×10-5 | |
| Christensenellaceae R7 group | IVW | 10 | 0.19 | 0.100 | 1.218 (1.001‒1.483) | 0.049 | True | 9.97×10-5 | |
| Coriobacteriales | IVW | 14 | -0.21 | 0.085 | 0.803 (0.679‒0.950) | 0.010 | True | 0.001 | |
| Coriobacteriia | IVW | 14 | -0.21 | 0.085 | 0.803 (0.679‒0.950) | 0.010 | True | 0.002 | |
| Desulfovibrio | IVW | 10 | 0.15 | 0.062 | 1.167 (1.033‒1.318) | 0.012 | True | 0.007 | |
| Coriobacteriaceae | IVW | 14 | -0.21 | 0.085 | 0.803 (0.679‒0.950) | 0.010 | true | 0.007 | |
| Intestinibacter | IVW | 15 | -0.19 | 0.083 | 0.819 (0.696‒0.965) | 0.010 | True | 0.015 | |
| HF | |||||||||
| Ruminococcaceae UCG009 | IVW | 12 | 0.06 | 0.030 | 1.107 (1.009‒1.137) | 0.022 | True | 6.12×10-6 | |
| Eubacterium oxidoreducens group | IVW | 4 | 0.11 | 0.043 | 1.117 (1.026‒1.215) | 0.010 | True | 3.26×10-5 | |
| Bacillales | IVW | 9 | -0.04 | 0.022 | 0.955 (0.913‒0.998) | 0.010 | True | 5.84×10-6 | |
| Selenomonadales | IVW | 12 | 0.10 | 0.044 | 1.106 (1.013‒1.208) | 0.023 | True | 1.53×10-5 | |
| Anaerostipes | IVW | 13 | -0.10 | 0.043 | 0.899 (0.825‒0.974) | 0.013 | True | 3.77×10-6 | |
| Negativicutes | IVW | 12 | 0.10 | 0.044 | 1.107 (1.014‒1.208) | 0.023 | True | 4.48×10-5 | |
| Eubacterium eligens group | IVW | 7 | 0.13 | 0.057 | 1.139 (1.019‒1.274) | 0.022 | True | 6.43×10-6 | |
| Flavonifractor | IVW | 5 | 0.13 | 0.053 | 1.144 (1.031‒1.270) | 0.011 | True | 1.06×10-5 |
表2 基于IVW法分析肠道微生物与CVD的因果关系
Tab 2 Causal relationship between gut microbiota and CVD based on IVW method
| Outcome | Exposure | MR method | SNP/n | β | se | OR (95%CI) | P value | Correct causal direction | P value (Steiger test) |
|---|---|---|---|---|---|---|---|---|---|
| AF | |||||||||
| Catenibacterium | IVW | 5 | 0.05 | 0.025 | 1.057 (1.005‒1.112) | 0.030 | True | 4.85×10-6 | |
| Victivallales | IVW | 8 | -0.06 | 0.023 | 0.939 (0.896‒0.984) | 0.008 | True | 1.52×10-6 | |
| Howardella | IVW | 10 | -0.06 | 0.019 | 0.939 (0.904‒0.977) | 0.001 | True | 3.04×10-5 | |
| Lachnospiraceae UCG008 | IVW | 11 | 0.04 | 0.025 | 1.051 (1.001‒1.104) | 0.047 | True | 2.52×10-7 | |
| Anaerostipes | IVW | 13 | -0.08 | 0.037 | 0.922 (0.857‒0.993) | 0.030 | True | 5.20×10-6 | |
| Bifidobacteriaceae | IVW | 11 | -0.08 | 0.034 | 0.916 (0.855‒0.980) | 0.011 | True | 1.42×10-5 | |
| Lentisphaeria | IVW | 8 | -0.06 | 0.024 | 0.936 (0.887‒0.983) | 0.003 | True | 3.96×10-6 | |
| Streptococcus | IVW | 15 | 0.08 | 0.039 | 1.089 (1.008‒1.177) | 0.029 | True | 9.37×10-6 | |
| Victivallis | IVW | 10 | 0.03 | 0.018 | 1.038 (1.001‒1.077) | 0.043 | True | 4.62×10-6 | |
| Odoribacter | IVW | 7 | -0.09 | 0.046 | 0.909 (0.831‒0.996) | 0.040 | True | 1.81×10-5 | |
| Intestinibacter | IVW | 15 | -0.06 | 0.031 | 0.933 (0.877‒0.993) | 0.028 | True | 2.28×10-6 | |
| Lentisphaerae | IVW | 9 | -0.07 | 0.022 | 0.926 (0.886‒0.968) | <0.001 | True | 5.06×10-6 | |
| Bifidobacteriales | IVW | 11 | -0.08 | 0.034 | 0.916 (0.855‒0.980) | 0.011 | True | 1.35×10-5 | |
| CAD | |||||||||
| Lactobacillales | IVW | 15 | -0.08 | 0.032 | 0.919 (0.862‒0.980) | 0.010 | True | 9.28×10-7 | |
| Veillonellaceae | IVW | 19 | 0.06 | 0.026 | 1.065 (1.011‒1.122) | 0.018 | True | 1.21×10-5 | |
| Parabacteroides | IVW | 6 | -0.14 | 0.050 | 0.866 (0.784‒0.956) | 0.004 | True | 6.31×10-5 | |
| Lachnospiraceae | IVW | 17 | 0.09 | 0.038 | 1.094 (1.012‒1.183) | 0.023 | True | 7.23×10-6 | |
| Lachnoclostridium | IVW | 13 | 0.08 | 0.039 | 1.093 (1.011‒1.181) | 0.025 | True | 9.14×10-5 | |
| Oxalobacter | IVW | 11 | 0.05 | 0.020 | 1.062 (1.019‒1.106) | 0.004 | True | 6.05×10-6 | |
| Odoribacter | IVW | 7 | 0.14 | 0.047 | 1.160 (1.056‒1.275) | 0.001 | True | 6.42×10-7 | |
| Hypertension | |||||||||
| Mollicutes RF9 | IVW | 13 | -0.16 | 0.068 | 0.851 (0.743‒0.974) | 0.019 | True | 7.23×10-5 | |
| Peptococcaceae | IVW | 9 | 0.20 | 0.086 | 1.230 (1.038‒1.458) | 0.016 | True | 2.07×10-5 | |
| Christensenellaceae R7 group | IVW | 10 | 0.19 | 0.100 | 1.218 (1.001‒1.483) | 0.049 | True | 9.97×10-5 | |
| Coriobacteriales | IVW | 14 | -0.21 | 0.085 | 0.803 (0.679‒0.950) | 0.010 | True | 0.001 | |
| Coriobacteriia | IVW | 14 | -0.21 | 0.085 | 0.803 (0.679‒0.950) | 0.010 | True | 0.002 | |
| Desulfovibrio | IVW | 10 | 0.15 | 0.062 | 1.167 (1.033‒1.318) | 0.012 | True | 0.007 | |
| Coriobacteriaceae | IVW | 14 | -0.21 | 0.085 | 0.803 (0.679‒0.950) | 0.010 | true | 0.007 | |
| Intestinibacter | IVW | 15 | -0.19 | 0.083 | 0.819 (0.696‒0.965) | 0.010 | True | 0.015 | |
| HF | |||||||||
| Ruminococcaceae UCG009 | IVW | 12 | 0.06 | 0.030 | 1.107 (1.009‒1.137) | 0.022 | True | 6.12×10-6 | |
| Eubacterium oxidoreducens group | IVW | 4 | 0.11 | 0.043 | 1.117 (1.026‒1.215) | 0.010 | True | 3.26×10-5 | |
| Bacillales | IVW | 9 | -0.04 | 0.022 | 0.955 (0.913‒0.998) | 0.010 | True | 5.84×10-6 | |
| Selenomonadales | IVW | 12 | 0.10 | 0.044 | 1.106 (1.013‒1.208) | 0.023 | True | 1.53×10-5 | |
| Anaerostipes | IVW | 13 | -0.10 | 0.043 | 0.899 (0.825‒0.974) | 0.013 | True | 3.77×10-6 | |
| Negativicutes | IVW | 12 | 0.10 | 0.044 | 1.107 (1.014‒1.208) | 0.023 | True | 4.48×10-5 | |
| Eubacterium eligens group | IVW | 7 | 0.13 | 0.057 | 1.139 (1.019‒1.274) | 0.022 | True | 6.43×10-6 | |
| Flavonifractor | IVW | 5 | 0.13 | 0.053 | 1.144 (1.031‒1.270) | 0.011 | True | 1.06×10-5 |
| Outcome | Exposure | Cochran's Q | MR-Egger intercept | |||||
|---|---|---|---|---|---|---|---|---|
| MR-Egger | IVW | Intercept | P value | |||||
| Q value | P value | Q value | P value | |||||
| AF | ||||||||
| Catenibacterium | 0.370 | 0.946 | 0.488 | 0.974 | 0.010 | 0.753 | ||
| Victivallales | 3.948 | 0.683 | 4.155 | 0.761 | 0.005 | 0.665 | ||
| Howardella | 7.364 | 0.497 | 7.622 | 0.572 | -0.006 | 0.624 | ||
| Lachnospiraceae UCG008 | 8.051 | 0.528 | 8.162 | 0.612 | 0.004 | 0.747 | ||
| Anaerostipes | 12.334 | 0.339 | 12.561 | 0.402 | -0.003 | 0.661 | ||
| Bifidobacteriaceae | 8.821 | 0.453 | 9.111 | 0.521 | 0.004 | 0.603 | ||
| Lentisphaeria | 3.948 | 0.683 | 4.155 | 0.761 | 0.005 | 0.665 | ||
| Streptococcus | 20.126 | 0.092 | 20.223 | 0.123 | 0.002 | 0.807 | ||
| Victivallis | 7.007 | 0.535 | 8.153 | 0.518 | 0.019 | 0.315 | ||
| Odoribacter | 3.621 | 0.605 | 4.634 | 0.591 | 0.010 | 0.360 | ||
| Intestinibacter | 14.962 | 0.309 | 17.041 | 0.253 | 0.010 | 0.203 | ||
| Lentisphaerae | 7.455 | 0.383 | 7.841 | 0.449 | 0.007 | 0.566 | ||
| Bifidobacteriales | 8.821 | 0.454 | 9.111 | 0.521 | 0.004 | 0.603 | ||
| CAD | ||||||||
| Lactobacillales | 13.551 | 0.406 | 14.344 | 0.424 | -0.005 | 0.398 | ||
| Veillonellaceae | 14.934 | 0.528 | 1.741 | 0.495 | 0.005 | 0.241 | ||
| Parabacteroides | 4.608 | 0.329 | 4.742 | 0.448 | 0.005 | 0.750 | ||
| Lachnospiraceae | 21.818 | 0.112 | 24.485 | 0.079 | -0.008 | 0.195 | ||
| Lachnoclostridium | 5.378 | 0.911 | 6.793 | 0.870 | 0.011 | 0.259 | ||
| Oxalobacter | 3.473 | 0.942 | 3.692 | 0.960 | -0.006 | 0.650 | ||
| Odoribacter | 4.080 | 0.537 | 5.288 | 0.507 | 0.012 | 0.321 | ||
| Hypertension | ||||||||
| Mollicutes RF9 | 3.784 | 0.975 | 0.711 | 0.877 | <0.001 | 0.957 | ||
| Peptococcaceae | 5.131 | 0.643 | 0.331 | 0.178 | 0.055 | 0.358 | ||
| Christensenellaceae R7 group | 6.448 | 0.597 | 7.960 | 0.538 | 0.008 | 0.575 | ||
| Coriobacteriales | 8.827 | 0.717 | 9.577 | 0.728 | 0.012 | 0.575 | ||
| Coriobacteriia | 8.827 | 0.717 | 9.577 | 0.728 | 0.013 | 0.145 | ||
| Desulfovibrio | 3.323 | 0.912 | 7.960 | 0.538 | 0.222 | 0.253 | ||
| Coriobacteriaceae | 8.827 | 0.717 | 9.577 | 0.728 | -0.020 | 0.403 | ||
| Intestinibacter | 19.114 | 0.119 | 21.572 | 0.087 | -0.020 | 0.403 | ||
| HF | ||||||||
| Ruminococcaceae UCG009 | 8.939 | 0.537 | 10.956 | 0.446 | 0.017 | 0.185 | ||
| Eubacterium oxidoreducens group | 2.451 | 0.293 | 2.483 | 0.478 | -0.002 | 0.887 | ||
| Bacillales | 2.705 | 0.910 | 2.870 | 0.942 | 0.006 | 0.697 | ||
| Selenomonadales | 4.459 | 0.924 | 5.180 | 0.922 | -0.007 | 0.415 | ||
| Anaerostipes | 9.973 | 0.532 | 10.057 | 0.610 | -0.002 | 0.777 | ||
| Negativicutes | 4.459 | 0.924 | 5.180 | 0.922 | -0.007 | 0.415 | ||
| Eubacterium eligens group | 1.547 | 0.907 | 6.428 | 0.376 | -0.038 | 0.078 | ||
| Flavonifractor | 2.919 | 0.404 | 2.919 | 0.404 | 0.002 | 0.903 | ||
表3 肠道微生物对CVD效应的异质性及水平多效性分析
Tab 3 Heterogeneity and horizontal pleiotropy analysis of the effect of gut microbiota on CVD
| Outcome | Exposure | Cochran's Q | MR-Egger intercept | |||||
|---|---|---|---|---|---|---|---|---|
| MR-Egger | IVW | Intercept | P value | |||||
| Q value | P value | Q value | P value | |||||
| AF | ||||||||
| Catenibacterium | 0.370 | 0.946 | 0.488 | 0.974 | 0.010 | 0.753 | ||
| Victivallales | 3.948 | 0.683 | 4.155 | 0.761 | 0.005 | 0.665 | ||
| Howardella | 7.364 | 0.497 | 7.622 | 0.572 | -0.006 | 0.624 | ||
| Lachnospiraceae UCG008 | 8.051 | 0.528 | 8.162 | 0.612 | 0.004 | 0.747 | ||
| Anaerostipes | 12.334 | 0.339 | 12.561 | 0.402 | -0.003 | 0.661 | ||
| Bifidobacteriaceae | 8.821 | 0.453 | 9.111 | 0.521 | 0.004 | 0.603 | ||
| Lentisphaeria | 3.948 | 0.683 | 4.155 | 0.761 | 0.005 | 0.665 | ||
| Streptococcus | 20.126 | 0.092 | 20.223 | 0.123 | 0.002 | 0.807 | ||
| Victivallis | 7.007 | 0.535 | 8.153 | 0.518 | 0.019 | 0.315 | ||
| Odoribacter | 3.621 | 0.605 | 4.634 | 0.591 | 0.010 | 0.360 | ||
| Intestinibacter | 14.962 | 0.309 | 17.041 | 0.253 | 0.010 | 0.203 | ||
| Lentisphaerae | 7.455 | 0.383 | 7.841 | 0.449 | 0.007 | 0.566 | ||
| Bifidobacteriales | 8.821 | 0.454 | 9.111 | 0.521 | 0.004 | 0.603 | ||
| CAD | ||||||||
| Lactobacillales | 13.551 | 0.406 | 14.344 | 0.424 | -0.005 | 0.398 | ||
| Veillonellaceae | 14.934 | 0.528 | 1.741 | 0.495 | 0.005 | 0.241 | ||
| Parabacteroides | 4.608 | 0.329 | 4.742 | 0.448 | 0.005 | 0.750 | ||
| Lachnospiraceae | 21.818 | 0.112 | 24.485 | 0.079 | -0.008 | 0.195 | ||
| Lachnoclostridium | 5.378 | 0.911 | 6.793 | 0.870 | 0.011 | 0.259 | ||
| Oxalobacter | 3.473 | 0.942 | 3.692 | 0.960 | -0.006 | 0.650 | ||
| Odoribacter | 4.080 | 0.537 | 5.288 | 0.507 | 0.012 | 0.321 | ||
| Hypertension | ||||||||
| Mollicutes RF9 | 3.784 | 0.975 | 0.711 | 0.877 | <0.001 | 0.957 | ||
| Peptococcaceae | 5.131 | 0.643 | 0.331 | 0.178 | 0.055 | 0.358 | ||
| Christensenellaceae R7 group | 6.448 | 0.597 | 7.960 | 0.538 | 0.008 | 0.575 | ||
| Coriobacteriales | 8.827 | 0.717 | 9.577 | 0.728 | 0.012 | 0.575 | ||
| Coriobacteriia | 8.827 | 0.717 | 9.577 | 0.728 | 0.013 | 0.145 | ||
| Desulfovibrio | 3.323 | 0.912 | 7.960 | 0.538 | 0.222 | 0.253 | ||
| Coriobacteriaceae | 8.827 | 0.717 | 9.577 | 0.728 | -0.020 | 0.403 | ||
| Intestinibacter | 19.114 | 0.119 | 21.572 | 0.087 | -0.020 | 0.403 | ||
| HF | ||||||||
| Ruminococcaceae UCG009 | 8.939 | 0.537 | 10.956 | 0.446 | 0.017 | 0.185 | ||
| Eubacterium oxidoreducens group | 2.451 | 0.293 | 2.483 | 0.478 | -0.002 | 0.887 | ||
| Bacillales | 2.705 | 0.910 | 2.870 | 0.942 | 0.006 | 0.697 | ||
| Selenomonadales | 4.459 | 0.924 | 5.180 | 0.922 | -0.007 | 0.415 | ||
| Anaerostipes | 9.973 | 0.532 | 10.057 | 0.610 | -0.002 | 0.777 | ||
| Negativicutes | 4.459 | 0.924 | 5.180 | 0.922 | -0.007 | 0.415 | ||
| Eubacterium eligens group | 1.547 | 0.907 | 6.428 | 0.376 | -0.038 | 0.078 | ||
| Flavonifractor | 2.919 | 0.404 | 2.919 | 0.404 | 0.002 | 0.903 | ||
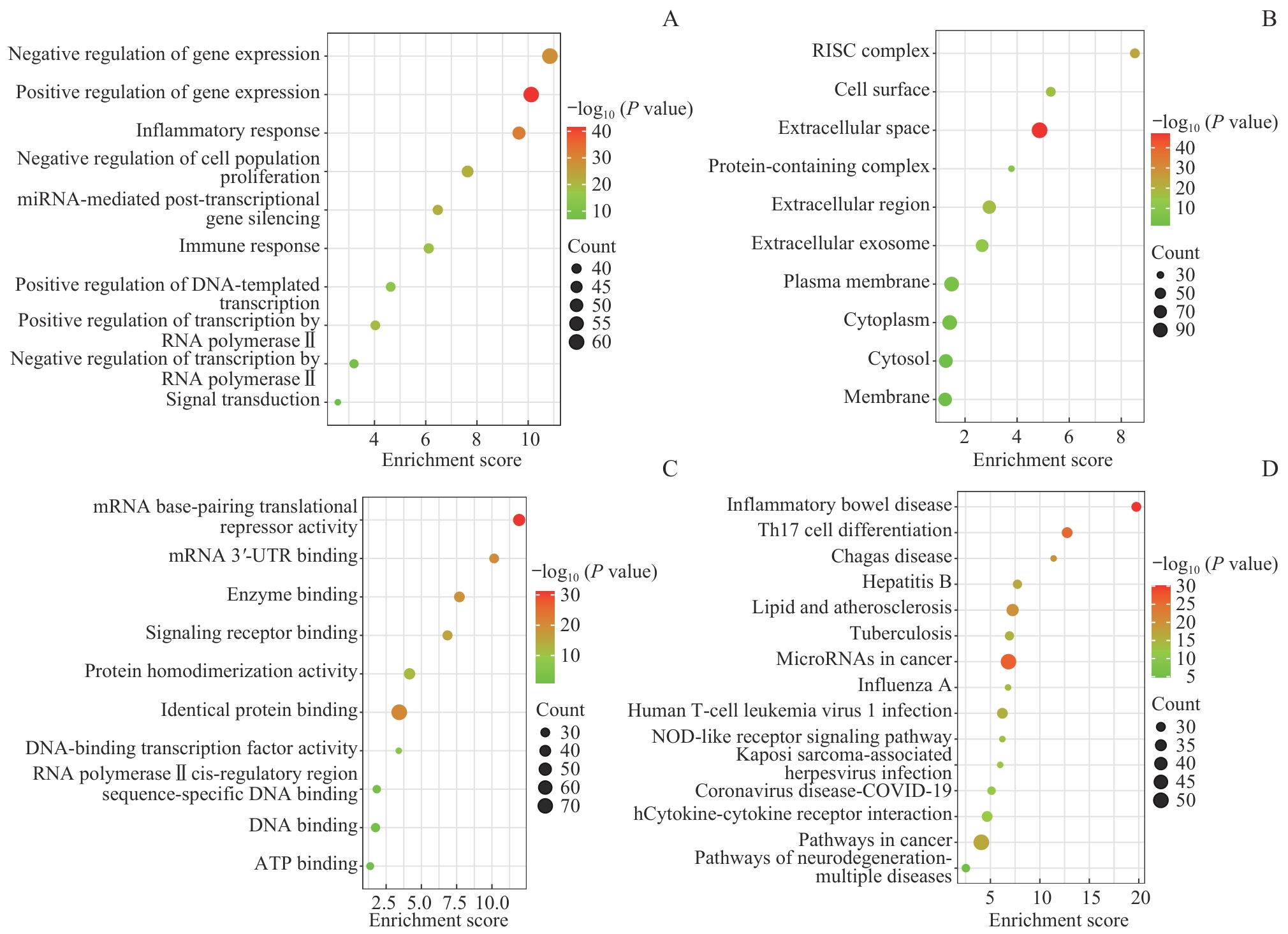
图5 影响CVD的肠道微生物群所携带基因的GO功能和KEGG通路富集分析Note: A. Biological process (GO). B. Cell component (GO). C. Molecular function (GO). D. KEGG analysis.
Fig 5 GO function and KEGG pathway enrichment analysis of genes carried by gut microbiota affecting CVD
| [1] | LI Y, CAO G Y, JING W Z, et al. Global trends and regional differences in incidence and mortality of cardiovascular disease, 1990‒2019: findings from 2019 global burden of disease study[J]. Eur J Prev Cardiol, 2023, 30(3): 276-286. |
| [2] | 刘明波, 何新叶, 杨晓红, 等. 《中国心血管健康与疾病报告2023》要点解读[J]. 临床心血管病杂志, 2024, 40(8): 599-616. |
| LIU M B, HE X Y, YANG X H, et al. Interpretation of Report on Cardiovascular Health and Diseases in China 2023[J]. Journal of Clinical Cardiology, 2024, 40(8): 599-616. | |
| [3] | SHI H Q, TER HORST R, NIELEN S, et al. The gut microbiome as mediator between diet and its impact on immune function[J]. Sci Rep, 2022, 12(1): 5149. |
| [4] | OJO O, FENG Q Q, OJO O O, et al. The role of dietary fibre in modulating gut microbiota dysbiosis in patients with type 2 diabetes: a systematic review and meta-analysis of randomised controlled trials[J]. Nutrients, 2020, 12(11): 3239. |
| [5] | TELLE-HANSEN V H, GAUNDAL L, BASTANI N, et al. Replacing saturated fatty acids with polyunsaturated fatty acids increases the abundance of Lachnospiraceae and is associated with reduced total cholesterol levels: a randomized controlled trial in healthy individuals[J]. Lipids Health Dis, 2022, 21(1): 92. |
| [6] | WANG Z N, KLIPFELL E, BENNETT B J, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease[J]. Nature, 2011, 472(7341): 57-63. |
| [7] | UFNAL M, JAZWIEC R, DADLEZ M, et al. Trimethylamine-N-oxide: a carnitine-derived metabolite that prolongs the hypertensive effect of angiotensin Ⅱ in rats[J]. Can J Cardiol, 2014, 30(12): 1700-1705. |
| [8] | LI D D, LU Y, YUAN S, et al. Gut microbiota-derived metabolite trimethylamine-N-oxide and multiple health outcomes: an umbrella review and updated meta-analysis[J]. Am J Clin Nutr, 2022, 116(1): 230-243. |
| [9] | BÄCKHED F, LEY R E, SONNENBURG J L, et al. Host-bacterial mutualism in the human intestine[J]. Science, 2005, 307(5717): 1915-1920. |
| [10] | SAVAGE D C. Associations of indigenous microorganisms with gastrointestinal mucosal epithelia[J]. Am J Clin Nutr, 1970, 23(11): 1495-1501. |
| [11] | MAZMANIAN S K, LIU C H, TZIANABOS A O, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system[J]. Cell, 2005, 122(1): 107-118. |
| [12] | LI M, WANG B H, ZHANG M H, et al. Symbiotic gut microbes modulate human metabolic phenotypes[J]. Proc Natl Acad Sci USA, 2008, 105(6): 2117-2122. |
| [13] | WANG H R, REN S J, LV H L, et al. Gut microbiota from mice with cerebral ischemia-reperfusion injury affects the brain in healthy mice[J]. Aging (Albany NY), 2021, 13(7): 10058-10074. |
| [14] | WAN S Z, NIE Y, ZHANG Y, et al. Gut microbial dysbiosis is associated with profibrotic factors in liver fibrosis mice[J]. Front Cell Infect Microbiol, 2020, 10: 18. |
| [15] | JIE Z Y, XIA H H, ZHONG S L, et al. The gut microbiome in atherosclerotic cardiovascular disease[J]. Nat Commun, 2017, 8(1): 845. |
| [16] | TANG W H W, BÄCKHED F, LANDMESSER U, et al. Intestinal microbiota in cardiovascular health and disease: JACC state-of-the-art review[J]. J Am Coll Cardiol, 2019, 73(16): 2089-2105. |
| [17] | WAN Y, WANG F L, YUAN J H, et al. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: a 6-month randomised controlled-feeding trial[J]. Gut, 2019, 68(8): 1417-1429. |
| [18] | DELANNOY-BRUNO O, DESAI C, RAMAN A S, et al. Evaluating microbiome-directed fibre snacks in gnotobiotic mice and humans[J]. Nature, 2021, 595(7865): 91-95. |
| [19] | SMITH G D, EBRAHIM S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease?[J]. Int J Epidemiol, 2003, 32(1): 1-22. |
| [20] | DAVEY SMITH G, HEMANI G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies[J]. Hum Mol Genet, 2014, 23(R1): R89-R98. |
| [21] | SEKULA P, DEL GRECO M F, PATTARO C, et al. Mendelian randomization as an approach to assess causality using observational data[J]. J Am Soc Nephrol, 2016, 27(11): 3253-3265. |
| [22] | WANG J, KURILSHIKOV A, RADJABZADEH D, et al. Meta-analysis of human genome-microbiome association studies: the MiBioGen consortium initiative[J]. Microbiome, 2018, 6(1): 101. |
| [23] | KURILSHIKOV A, MEDINA-GOMEZ C, BACIGALUPE R, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition[J]. Nat Genet, 2021, 53(2): 156-165. |
| [24] | WU F S, HUANG Y, HU J L, et al. Mendelian randomization study of inflammatory bowel disease and bone mineral density[J]. BMC Med, 2020, 18(1): 312. |
| [25] | BURGESS S, BUTTERWORTH A, THOMPSON S G. Mendelian randomization analysis with multiple genetic variants using summarized data[J]. Genet Epidemiol, 2013, 37(7): 658-665. |
| [26] | LONG Y W, TANG L H, ZHOU Y Y, et al. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study[J]. BMC Med, 2023, 21(1): 66. |
| [27] | LAWLOR D A, HARBORD R M, STERNE J A C, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology[J]. Stat Med, 2008, 27(8): 1133-1163. |
| [28] | HEMANI G, TILLING K, DAVEY SMITH G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data[J]. PLoS Genet, 2017, 13(11): e1007081. |
| [29] | TANG W H, WANG Z N, LEVISON B S, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk[J]. N Engl J Med, 2013, 368(17): 1575-1584. |
| [30] | KOH A, DE VADDER F, KOVATCHEVA-DATCHARY P, et al. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites[J]. Cell, 2016, 165(6): 1332-1345. |
| [31] | WAHLSTRÖM A, SAYIN S I, MARSCHALL H U, et al. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism[J]. Cell Metab, 2016, 24(1): 41-50. |
| [32] | GUO Y, LUO S Y, YE Y X, et al. Intermittent fasting improves cardiometabolic risk factors and alters gut microbiota in metabolic syndrome patients[J]. J Clin Endocrinol Metab, 2021, 106(1): 64-79. |
| [33] | KOREN O, SPOR A, FELIN J, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis[J]. Proc Natl Acad Sci USA, 2011, 108(Suppl 1): 4592-4598. |
| [34] | HUTCHINSON A N, TINGÖ L N, BRUMMER R J. The potential effects of probiotics and ω-3 fatty acids on chronic low-grade inflammation[J]. Nutrients, 2020, 12(8): 2402. |
| [35] | BRON P A, KLEEREBEZEM M, BRUMMER R J, et al. Can probiotics modulate human disease by impacting intestinal barrier function?[J]. Br J Nutr, 2017, 117(1): 93-107. |
| [36] | RUSCICA M, PAVANELLO C, GANDINI S, et al. Nutraceutical approach for the management of cardiovascular risk—a combination containing the probiotic Bifidobacterium longum BB536 and red yeast rice extract: results from a randomized, double-blind, placebo-controlled study[J]. Nutr J, 2019, 18(1): 13. |
| [37] | LIU J L, AN N, MA C, et al. Correlation analysis of intestinal flora with hypertension[J]. Exp Ther Med, 2018, 16(3): 2325-2330. |
| [38] | MAYERHOFER C C K, KUMMEN M, HOLM K, et al. Low fibre intake is associated with gut microbiota alterations in chronic heart failure[J]. ESC Heart Fail, 2020, 7(2): 456-466. |
| [39] | WANG M, XIONG H, LU L, et al. Serum lipopolysaccharide is associated with the recurrence of atrial fibrillation after radiofrequency ablation by increasing systemic inflammation and atrial fibrosis[J]. Oxid Med Cell Longev, 2022, 2022: 2405972. |
| [40] | KONG B, FU H, XIAO Z, et al. Gut microbiota dysbiosis induced by a high-fat diet increases susceptibility to atrial fibrillation[J]. Can J Cardiol, 2022, 38(12): 1962-1975. |
| [41] | INZAUGARAT M E, JOHNSON C D, HOLTMANN T M, et al. NLR family pyrin domain-containing 3 inflammasome activation in hepatic stellate cells induces liver fibrosis in mice[J]. Hepatology, 2019, 69(2): 845-859. |
| [42] | LIU P N, YU S S, LIU J R, et al. Effects of Lactobacillus on hyperlipidemia in high-fat diet-induced mouse model[J]. Arch Med Sci, 2020, 19(3): 792-799. |
| [43] | CHEN L H, LIU W E, LI Y M, et al. Lactobacillus acidophilus ATCC 4356 attenuates the atherosclerotic progression through modulation of oxidative stress and inflammatory process[J]. Int Immunopharmacol, 2013, 17(1): 108-115. |
| [44] | HUANG Y, WANG J F, QUAN G H, et al. Lactobacillus acidophilus ATCC 4356 prevents atherosclerosis via inhibition of intestinal cholesterol absorption in apolipoprotein E-knockout mice[J]. Appl Environ Microbiol, 2014, 80(24): 7496-7504. |
| [45] | AHN H Y, KIM M, CHAE J S, et al. Supplementation with two probiotic strains, Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032, reduces fasting triglycerides and enhances apolipoprotein A-V levels in non-diabetic subjects with hypertriglyceridemia[J]. Atherosclerosis, 2015, 241(2): 649-656. |
| [46] | 李亚梦, 崔美泽, 孙婧, 等. 肠道菌群及其代谢产物氧化三甲胺: 冠心病治疗的新靶点[J]. 生物工程学报, 2021, 37(11): 3745-3756. |
| LI Y M, CUI M Z, SUN J, et al. Gut microbiota and its metabolite trimethylamine-N-oxide (TMAO): a novel regulator in coronary artery disease[J]. Chinese Journal of Biotechnology, 2021, 37(11): 3745-3756. | |
| [47] | KATSIMICHAS T, THEOFILIS P, TSIOUFIS K, et al. Gut microbiota and coronary artery disease: current therapeutic perspectives[J]. Metabolites, 2023, 13(2): 256. |
| [48] | WANG Y, XU Y Y, YANG M, et al. Butyrate mitigates TNF-α-induced attachment of monocytes to endothelial cells[J]. J Bioenerg Biomembr, 2020, 52(4): 247-256. |
| [49] | CHEN W J, ZHANG S, WU J F, et al. Butyrate-producing bacteria and the gut-heart axis in atherosclerosis[J]. Clin Chim Acta, 2020, 507: 236-241. |
| [50] | YU Y J, RAKA F, ADELI K. The role of the gut microbiota in lipid and lipoprotein metabolism[J]. J Clin Med, 2019, 8(12): 2227. |
| [51] | TORTELOTE G G. Therapeutic strategies for hypertension: exploring the role of microbiota-derived short-chain fatty acids in kidney physiology and development[J]. Pediatr Nephrol, 2025. |
| [52] | LUO Q, HU Y L, CHEN X, et al. Effects of gut microbiota and metabolites on heart failure and its risk factors: a two-sample Mendelian randomization study[J]. Front Nutr, 2022, 9: 899746. |
| [53] | CHIONCEL O, AMBROSY A P. Trimethylamine N-oxide and risk of heart failure progression: marker or mediator of disease[J]. Eur J Heart Fail, 2019, 21(7): 887-890. |
| [54] | SUN X L, JIAO X F, MA Y R, et al. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome[J]. Biochem Biophys Res Commun, 2016, 481(1/2): 63-70. |
| [55] | KEITEL V, REINEHR R, GATSIOS P, et al. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells[J]. Hepatology, 2007, 45(3): 695-704. |
| [56] | ROMANO K A, MARTINEZ-DEL CAMPO A, KASAHARA K, et al. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption[J]. Cell Host Microbe, 2017, 22(3): 279-290.e7. |
| [1] | 李龙, 赵霞, 金珊, 李泽莹, 吕福强, 庞丽娟, 刘克坚. 孟德尔随机化解析AZGP1在心力衰竭中的保护作用[J]. 上海交通大学学报(医学版), 2025, 45(8): 1035-1045. |
| [2] | 闫军浩, 郭晓磊, 罗昭锋, 唐坚, 王争. 孟德尔随机化分析乳糜泻与自身免疫性甲状腺疾病的因果关系[J]. 上海交通大学学报(医学版), 2025, 45(6): 766-773. |
| [3] | 徐苓, 皇甫昱婵, 沈立松, 马妍慧. 两样本孟德尔随机化分析血浆磷脂酰乙醇胺水平与结直肠腺癌发病风险的关系[J]. 上海交通大学学报(医学版), 2025, 45(5): 605-613. |
| [4] | 张慧华, 干静, 侯媌媌, 卢娜. 阻塞性睡眠呼吸暂停与脑成像衍生表型的双向孟德尔随机化研究[J]. 上海交通大学学报(医学版), 2025, 45(4): 468-475. |
| [5] | 林祎嘉, 程丽珍, 胡廷军, 苗雅. 基于孟德尔随机化法的2型糖尿病与认知障碍因果关系研究[J]. 上海交通大学学报(医学版), 2025, 45(2): 204-210. |
| [6] | 张迎迎, 张俊瑶, 宋际伟, 王声杰, 姚俊岩. 空气污染与阿尔茨海默病因果关联的两样本孟德尔随机化研究[J]. 上海交通大学学报(医学版), 2025, 45(1): 87-94. |
| [7] | 俞洋, 孟丹, 仇奕文, 袁见, 朱莹杰. 两样本孟德尔随机化法分析1型糖尿病对结直肠癌的影响[J]. 上海交通大学学报(医学版), 2024, 44(6): 755-761. |
| [8] | 李萍, 蒋惠如, 叶梦月, 王雅玉, 陈潇雨, 袁安彩, 徐文杰, 戴慧敏, 陈曦, 闫小响, 涂圣贤, 郑元琦, 张薇, 卜军. 基于上海社区老年人群队列的心血管疾病和恶性肿瘤的危险因素流行特征分析[J]. 上海交通大学学报(医学版), 2024, 44(5): 617-625. |
| [9] | 高雄, 张秋霞, 杨苗苗, 罗玮, 王月刚, 修建成. 房颤与认知障碍的因果关系:一项孟德尔随机化研究[J]. 上海交通大学学报(医学版), 2023, 43(11): 1359-1365. |
| [10] | 何志洁, 何津春, 张燕培, 王耀东, 王占科. 基于垂直密度梯度离心全自动血脂谱检测法的家族性高三酰甘油血症家系血脂亚组分分析[J]. 上海交通大学学报(医学版), 2022, 42(4): 482-489. |
| [11] | 张彤, 田雪, 左颖婷, 郑曼琪, 张怡君, 吴寿岭, 陈朔华, 马高亭, 佟旭, 王安心, 莫大鹏. 无传统危险因素人群中TyG指数与心脑血管疾病的关系[J]. 上海交通大学学报(医学版), 2022, 42(3): 267-274. |
| [12] | 王昊, 王然, 巴乾. 食品中二氧化钛纳米材料对消化道组织及肠道微生物群影响的研究进展[J]. 上海交通大学学报(医学版), 2022, 42(2): 225-229. |
| [13] | 张静静, 祝超瑜, 肖元元, 蒋伏松, 高清歌, 方云云, 魏丽. 胰高血糖素样肽1受体基因rs3765467变异与2型糖尿病的关联研究[J]. 上海交通大学学报(医学版), 2021, 41(9): 1215-1221. |
| [14] | 张瀛丹, 王振. 肠道微生物群在强迫症发病机制及治疗中的作用研究进展[J]. 上海交通大学学报(医学版), 2021, 41(7): 967-971. |
| [15] | 蔡明琪, 陈焱, 林开斌, 黄冬. 生长分化因子11在心血管疾病中的作用[J]. 上海交通大学学报(医学版), 2021, 41(6): 834-838. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||