
上海交通大学学报(医学版) ›› 2024, Vol. 44 ›› Issue (2): 169-182.doi: 10.3969/j.issn.1674-8115.2024.02.003
司春婴1,2( ), 王建茹2, 李晓辉2(
), 王建茹2, 李晓辉2( ), 王永霞1,2, 关怀敏2
), 王永霞1,2, 关怀敏2
收稿日期:2023-05-19
接受日期:2023-12-11
出版日期:2024-02-28
发布日期:2024-02-28
通讯作者:
李晓辉,电子信箱:478103511@qq.com。作者简介:司春婴(1987—),男,讲师,主治医师,硕士;电子信箱:chunyingsi1987@163.com。
基金资助:
SI Chunying1,2( ), WANG Jianru2, LI Xiaohui2(
), WANG Jianru2, LI Xiaohui2( ), WANG Yongxia1,2, GUAN Huaimin2
), WANG Yongxia1,2, GUAN Huaimin2
Received:2023-05-19
Accepted:2023-12-11
Online:2024-02-28
Published:2024-02-28
Contact:
LI Xiaohui,E-mail:478103511@qq.com.Supported by:摘要:
目的·利用单细胞RNA测序技术(single-cell?RNA?sequencing,scRNA-Seq)阐释冠状动脉粥样硬化(coronary atherosclerosis,CA)的细胞通信景观,挖掘主导细胞亚群及其关键基因。方法·下载GSE131778数据集并进行预处理、质控、降维聚类及注释;利用CellChat包进行细胞通信分析,识别主导细胞亚群。利用FindAllMarker函数,筛选主导细胞亚群与其他细胞亚群间的差异表达基因(differentially expressed genes,DEGs),并构建其蛋白相互作用(protein-protein interaction,PPI)网络,将Degree算法排序前五位的DEGs作为关键基因。将关键基因与CellChat分析出的细胞通信网络进行匹配和挖掘,获取关键基因参与的配体-受体对(ligand-receptor pair,L-R)及其介导的信号通路,并对结果进行可视化。构建动脉粥样硬化小鼠模型,并利用反转录聚合酶链式反应检测关键基因在颈动脉粥样硬化病变处的表达情况。结果·在CA病变处共鉴定出11个细胞亚群,包括平滑肌细胞、内皮细胞、巨噬细胞、单核细胞等。细胞通信分析结果显示,CellChat在11个细胞亚群中检测到70对显著的L-R和26条相关的信号通路;平滑肌细胞处于通信活跃状态,与其他细胞亚群间相互作用的次数和强度最显著,是主导细胞亚群。DEGs筛选结果显示,平滑肌细胞亚群和其他细胞亚群之间共有206个DEGs,其中ITGB2、PTPRC、CCL2、DCN、IGF1被识别为关键基因。关键基因介导的细胞通信分析结果显示:CCL2与ACKR1形成L-R,通过介导CCL信号通路参与平滑肌细胞与内皮细胞间的通信网络;ITGB2分别与ITGAM、ITGAX组成受体复合物,再与C3形成L-R介导补体信号通路,参与平滑肌细胞与巨噬细胞、单核细胞间的通信网络。动物实验对关键基因的验证结果同生物信息学分析的结果一致。结论·平滑肌细胞在CA病理过程中是主导细胞,与其他细胞间有广泛的通信网络,可通过CCL2-ACKR1、C3-(ITGAM+ITGB2)和C3-(ITGAX+ITGB2)介导的CCL和补体信号通路,与内皮细胞、巨噬细胞和单核细胞构建细胞通信网络。
中图分类号:
司春婴, 王建茹, 李晓辉, 王永霞, 关怀敏. 基于单细胞测序技术解析冠状动脉粥样硬化患者平滑肌细胞的细胞间通信及关键基因[J]. 上海交通大学学报(医学版), 2024, 44(2): 169-182.
SI Chunying, WANG Jianru, LI Xiaohui, WANG Yongxia, GUAN Huaimin. Study on intercellular communication and key genes of smooth muscle cells in human coronary atherosclerosis based on single cell sequencing technology[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(2): 169-182.
| Primer | Sequence (5'→3') | Length/bp |
|---|---|---|
| ITGB2-F | ACCTCATGGATCTCTCCTACTC | 274 |
| ITGB2-R | CCGACCTCTGTCTGAAACTG | |
| PTPRC-F | AGTCTCTACGCAAAGCACGG | 254 |
| PTPRC-R | AGCACTATTGGTAGGCTCCG | |
| CCL2-F | GCTGACCCCAAGAAGGAATG | 183 |
| CCL2-R | TGAGGTGGTTGTGGAAAAGG | |
| DCN-F | AATCCCTTATGACCCTGACA | 275 |
| DCN-R | TTTCCAACTTCACGAGAGGT | |
| IGF1-F | GATGCTCTTCAGTTCGTGTG | 259 |
| IGF1-R | GCTTCGTTTTCTTGTTTGTC | |
| GAPDH-F | CCTTCCGTGTTCCTAC | 152 |
| GAPDH-R | GACAACCTGGTCCTCA |
表1 RT-PCR引物序列
Tab 1 Primer sequence for RT-PCR
| Primer | Sequence (5'→3') | Length/bp |
|---|---|---|
| ITGB2-F | ACCTCATGGATCTCTCCTACTC | 274 |
| ITGB2-R | CCGACCTCTGTCTGAAACTG | |
| PTPRC-F | AGTCTCTACGCAAAGCACGG | 254 |
| PTPRC-R | AGCACTATTGGTAGGCTCCG | |
| CCL2-F | GCTGACCCCAAGAAGGAATG | 183 |
| CCL2-R | TGAGGTGGTTGTGGAAAAGG | |
| DCN-F | AATCCCTTATGACCCTGACA | 275 |
| DCN-R | TTTCCAACTTCACGAGAGGT | |
| IGF1-F | GATGCTCTTCAGTTCGTGTG | 259 |
| IGF1-R | GCTTCGTTTTCTTGTTTGTC | |
| GAPDH-F | CCTTCCGTGTTCCTAC | 152 |
| GAPDH-R | GACAACCTGGTCCTCA |
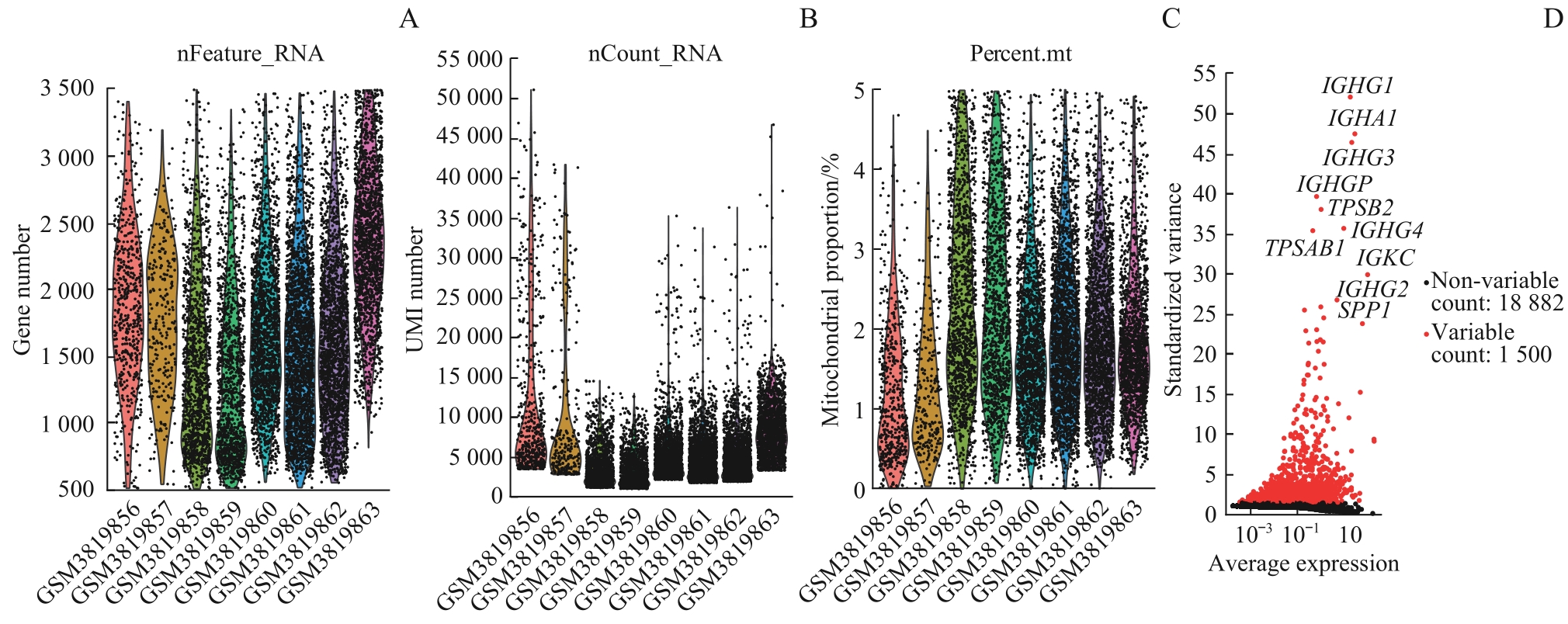
图1 GSE131778数据集样本的质控结果Note:A. Number of cell genes in the sample. B. Number of transcript sequencing counts in the sample. C. The proportion of mitochondria in all the cells in the sample. D. The top 1 500 mutated genes with high intercellular standard deviation.
Fig1 Quality control results for samples from the GSE131778 dataset

图2 GSE131778数据集中样本的降维聚类及注释结果Note:A.The distribution of cells in the sample in PC1 and PC2. B. P value for each PC. C. t-SNE plot of clustering distribution of 18 cell subpopulations. D. t-SNE diagram of the distribution of cell subpopulations after annotation. NK cell—nature killer cell; DC—dendritic cell.
Fig 2 Dimension reduction clustering and annotation results for samples in the GSE131778 dataset
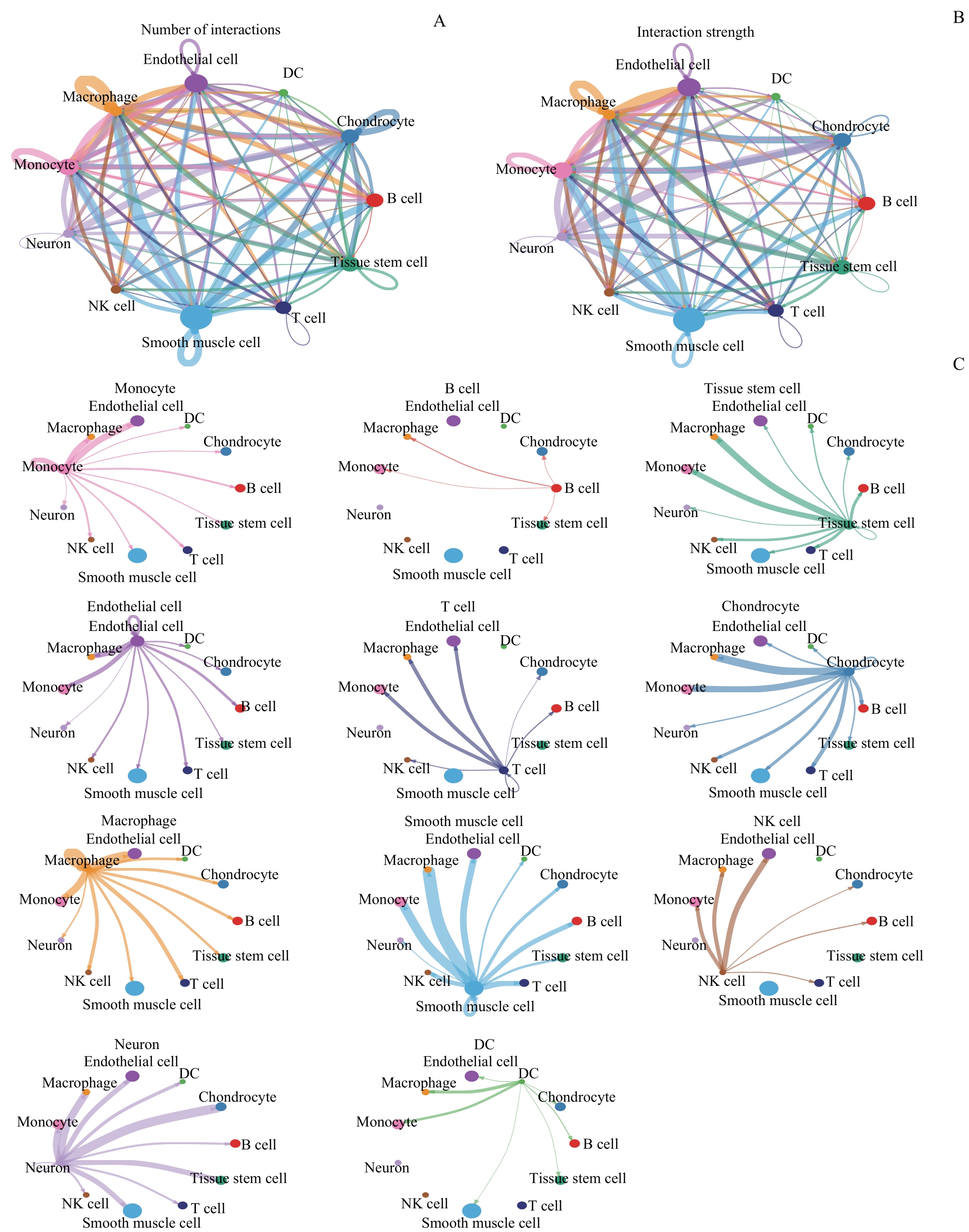
图3 CA病变处的细胞通信网络Note:A.Communication network diagram of the number of interactions among different cell clusters. B.Communication network diagram of interaction strength among different cell clusters. C. Communication network diagram of the interaction strength between a single cell cluster and other clusters. The color of the dots represents different cell clusters. The size of the dots represents the number of cells contained in the cell cluster, and the larger the dots, the more cells there are. The line represents the interaction relationship among the clusters, and the color represents the signal sent from the cluster as the sender to the cluster as the receiver. The thickness of the line represents the number of interactions (strength) among the clusters, and the thicker the line, the more interactions (strength) there are.
Fig 3 Cellular communication network at CA lesions

图4 平滑肌细胞DEGs的筛选及其富集分析Note:A.Volcano plot of DEGs in smooth muscle cells (dots represent genes, black represents genes with no differential expression, red represents upregulated DEGs, and blue represents downregulated DEGs). B. Bubble plots for GO enrichment analysis of DEGs. C. Circle diagram of KEGG enrichment analysis of DEGs.
Fig 4 Screening of DEGs in smooth muscle cells and their enrichment analysis

图5 平滑肌细胞关键基因的筛选结果Note: A. PPI network diagram of smooth muscle cell DEGs, with red circles representing upregulated DEGs and green triangles representing downregulated DEGs. B. PPI diagram of 5 hub genes. C. Circle diagram of chromosomal positions of hub genes. D. Bubble plot of 5 hub genes expressed in 11 clusters.
Fig 5 Results of screening for hub genes in smooth muscle cells
| Source | Target | Ligand | Receptor | Probability | P value | Interaction name | Pathway | Annotation | Evidence (PMID) |
|---|---|---|---|---|---|---|---|---|---|
| Chondrocyte | Endothelial cell | CCL2 | ACKR1 | 0.053 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Macrophage | Endothelial cell | CCL2 | ACKR1 | 0.042 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Monocyte | Endothelial cell | CCL2 | ACKR1 | 0.047 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Neuron | Endothelial cell | CCL2 | ACKR1 | 0.060 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Smooth muscle cell | Endothelial cell | CCL2 | ACKR1 | 0.244 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Tissue stem cell | Endothelial cell | CCL2 | ACKR1 | 0.021 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Macrophage | Macrophage | C3 | ITGAM_ITGB2 | 0.004 | 0.000 | C3-(ITGAM+ITGB2) | Complement | Secreted signaling | 16234578 |
| Smooth muscle cell | Macrophage | C3 | ITGAM_ITGB2 | 0.040 | 0.000 | C3-(ITGAM+ITGB2) | Complement | Secreted signaling | 16234578 |
| Macrophage | Monocyte | C3 | ITGAM_ITGB2 | 0.005 | 0.000 | C3-(ITGAM+ITGB2) | Complement | Secreted signaling | 16234578 |
| Smooth muscle cell | Monocyte | C3 | ITGAM_ITGB2 | 0.046 | 0.000 | C3-(ITGAM+ITGB2) | Complement | Secreted signaling | 16234578 |
| Macrophage | Macrophage | C3 | ITGAX_ITGB2 | 0.004 | 0.000 | C3-(ITGAX+ITGB2) | Complement | Secreted signaling | 16234578 |
| Smooth muscle cell | Macrophage | C3 | ITGAX_ITGB2 | 0.040 | 0.000 | C3-(ITGAX+ITGB2) | Complement | Secreted signaling | 16234578 |
表2 关键基因介导的细胞间信号通路
Tab 2 Hub genes-mediated cell signaling pathway
| Source | Target | Ligand | Receptor | Probability | P value | Interaction name | Pathway | Annotation | Evidence (PMID) |
|---|---|---|---|---|---|---|---|---|---|
| Chondrocyte | Endothelial cell | CCL2 | ACKR1 | 0.053 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Macrophage | Endothelial cell | CCL2 | ACKR1 | 0.042 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Monocyte | Endothelial cell | CCL2 | ACKR1 | 0.047 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Neuron | Endothelial cell | CCL2 | ACKR1 | 0.060 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Smooth muscle cell | Endothelial cell | CCL2 | ACKR1 | 0.244 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Tissue stem cell | Endothelial cell | CCL2 | ACKR1 | 0.021 | 0.000 | CCL2-ACKR1 | CCL | Secreted signaling | 26740381 |
| Macrophage | Macrophage | C3 | ITGAM_ITGB2 | 0.004 | 0.000 | C3-(ITGAM+ITGB2) | Complement | Secreted signaling | 16234578 |
| Smooth muscle cell | Macrophage | C3 | ITGAM_ITGB2 | 0.040 | 0.000 | C3-(ITGAM+ITGB2) | Complement | Secreted signaling | 16234578 |
| Macrophage | Monocyte | C3 | ITGAM_ITGB2 | 0.005 | 0.000 | C3-(ITGAM+ITGB2) | Complement | Secreted signaling | 16234578 |
| Smooth muscle cell | Monocyte | C3 | ITGAM_ITGB2 | 0.046 | 0.000 | C3-(ITGAM+ITGB2) | Complement | Secreted signaling | 16234578 |
| Macrophage | Macrophage | C3 | ITGAX_ITGB2 | 0.004 | 0.000 | C3-(ITGAX+ITGB2) | Complement | Secreted signaling | 16234578 |
| Smooth muscle cell | Macrophage | C3 | ITGAX_ITGB2 | 0.040 | 0.000 | C3-(ITGAX+ITGB2) | Complement | Secreted signaling | 16234578 |
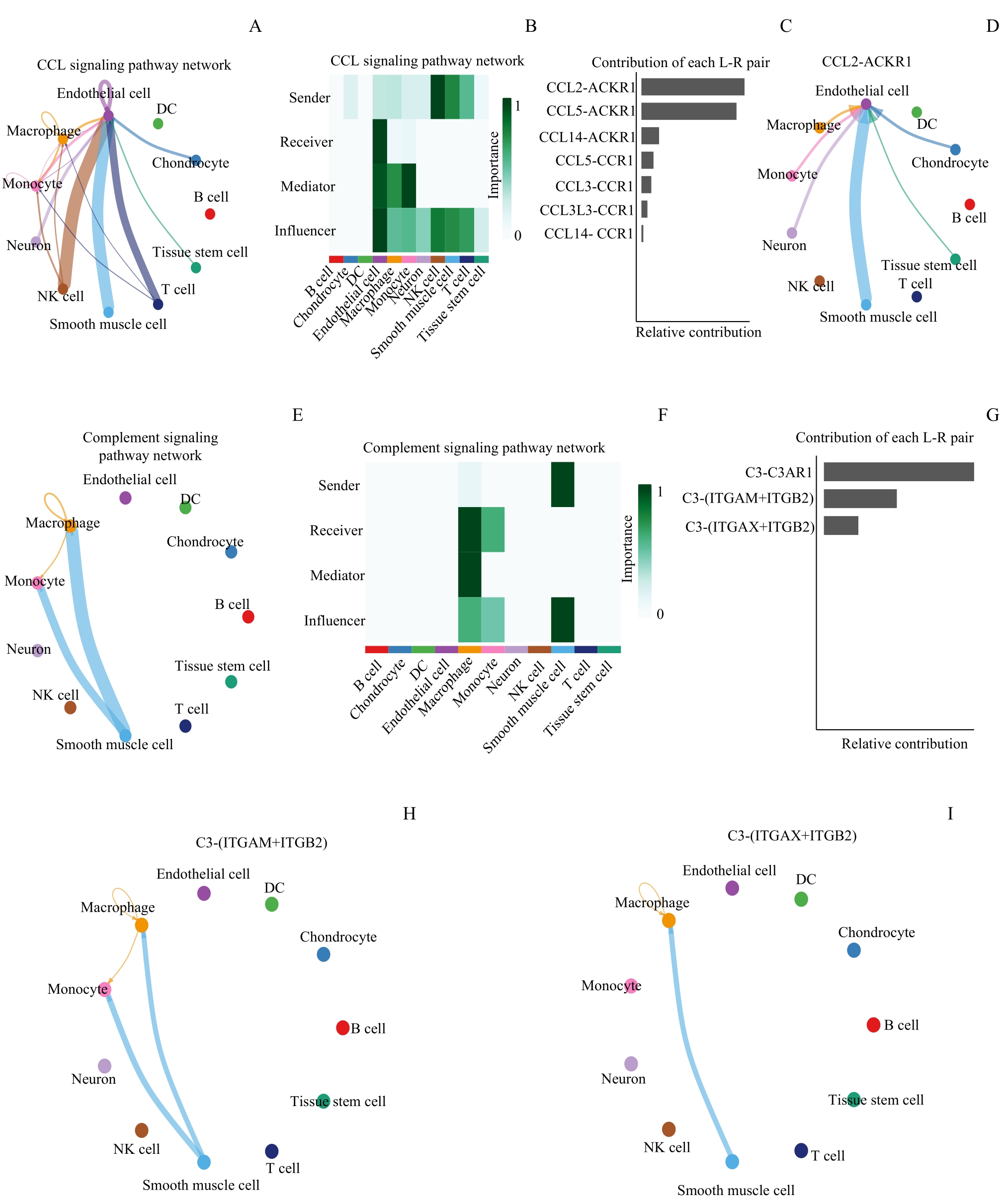
图6 关键基因介导的细胞间通信Note: A. Cellular communication network diagram mediated by CCL signaling pathway. B. Heat map of cell action types in the CCL signaling pathway. C. Bar chart of the contribution of L-R in the CCL signaling pathway. D. CCL2-ACKR1-mediated cellular communication network diagram. E. Cellular communication network diagram mediated by complement signaling pathway. F. Heat map of cellular action types in complement signaling pathways. G. Bar graph of the contribution of L-R in the complement signaling pathway. H. C3-(ITGAM+ITGB2)-mediated cellular communication network diagram. I. C3-(ITGAX+ITGB2)-mediated cellular communication network diagram.
Fig 6 Hub genes-mediated cellular communication
| Group | ITGB2 | PTPRC | CCL2 | DCN | IGF1 |
|---|---|---|---|---|---|
| Control | 1.04±0.34 | 1.00±0.06 | 1.01±0.19 | 1.01±0.15 | 1.00±0.06 |
| AS | 0.45±0.11① | 0.21±0.01② | 2.19±0.57③ | 1.57±0.11④ | 4.24±1.14⑤ |
表3 各组小鼠关键基因mRNA表达水平 (x±s, n=3)
Tab 3 mRNA expression levels of hub genes in each group of mice (x±s, n=3)
| Group | ITGB2 | PTPRC | CCL2 | DCN | IGF1 |
|---|---|---|---|---|---|
| Control | 1.04±0.34 | 1.00±0.06 | 1.01±0.19 | 1.01±0.15 | 1.00±0.06 |
| AS | 0.45±0.11① | 0.21±0.01② | 2.19±0.57③ | 1.57±0.11④ | 4.24±1.14⑤ |
| 1 | 郝俊海, 林展翼. 冠状动脉粥样硬化相关生物力学因素的研究进展[J]. 中国动脉硬化杂志, 2020, 28(11): 1009-1012. |
| HAO J H, LIN Z Y. Research progress of biomechanical factors related to coronary atherosclerosis[J]. Chinese Journal of Arteriosclerosis, 2020, 28(11): 1009-1012. | |
| 2 | HOSEN M R, GOODY P R, ZIETZER A, et al. MicroRNAs as master regulators of atherosclerosis: from pathogenesis to novel therapeutic options[J]. Antioxid Redox Signal, 2020, 33(9): 621-644. |
| 3 | D'ASCENZO F, AGOSTONI P, ABBATE A, et al. Atherosclerotic coronary plaque regression and the risk of adverse cardiovascular events: a meta-regression of randomized clinical trials[J]. Atherosclerosis, 2013, 226(1): 178-185. |
| 4 | BERUMEN SÁNCHEZ G, BUNN K E, PUA H H, et al. Extracellular vesicles: mediators of intercellular communication in tissue injury and disease[J]. Cell Commun Signal, 2021, 19(1): 104. |
| 5 | CHARLA E, MERCER J, MAFFIA P, et al. Extracellular vesicle signalling in atherosclerosis[J]. Cell Signal, 2020, 75: 109751. |
| 6 | ZHANG L Z, LEI S. Changes of junctions of endothelial cells in coronary sclerosis: a review[J]. Chronic Dis Transl Med, 2016, 2(1): 22-26. |
| 7 | WEN D, WANG X, CHEN R, et al. Single-cell RNA sequencing reveals the pathogenic relevance of intracranial atherosclerosis in blood blister-like aneurysms[J]. Front Immunol, 2022, 13: 927125. |
| 8 | JIN S, RAMOS R. Computational exploration of cellular communication in skin from emerging single-cell and spatial transcriptomic data[J]. Biochem Soc Trans, 2022, 50(1): 297-308. |
| 9 | POTTER S S. Single-cell RNA sequencing for the study of development, physiology and disease[J]. Nat Rev Nephrol, 2018, 14(8): 479-492. |
| 10 | COCHAIN C, VAFADARNEJAD E, ARAMPATZI P, et al. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis[J]. Circ Res, 2018, 122(12): 1661-1674. |
| 11 | WIRKA R C, WAGH D, PAIK D T, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis[J]. Nat Med, 2019, 25(8): 1280-1289. |
| 12 | TILLIE R J H A, VAN KUIJK K, SLUIMER J C. Fibroblasts in atherosclerosis: heterogeneous and plastic participants[J]. Curr Opin Lipidol, 2020, 31(5): 273-278. |
| 13 | 王建茹, 李晓辉. 基于单细胞RNA测序技术筛选颈动脉粥样硬化中巨噬细胞特征基因的研究[J]. 医学研究生学报, 2022, 35(10):1014-1021. |
| WANG J R, LI X H. Screening of macrophage characteristic genes in carotid atherosclerosis by single-cell RNA sequencing[J]. Journal of Medical Graduate students, 2022, 35(10): 1014-1021. | |
| 14 | JIN S, GUERRERO-JUAREZ C F, ZHANG L, et al. Inference and analysis of cell-cell communication using CellChat[J]. Nat Commun, 2021, 12(1): 1088. |
| 15 | 王建茹, 朱明军, 王永霞, 等. 基于网络药理学和分子对接技术探讨芪参益气滴丸改善心肌缺血再灌注损伤的潜在分子机制[J]. 中医学报, 2021, 36(7): 1537-1544. |
| WANG J R, ZHU M J, WANG Y X, et al. Study on the potential molecular mechanism of Qishenyiqi Dropping pills to improve myocardial ischemia reperfusion injury based on network pharmacology and molecular docking technique[J]. Journal of Traditional Chinese Medicine, 2021, 36(7): 1537-1544. | |
| 16 | 陈馨浓, 葛其卉, 赵一璇, 等. 四妙勇安汤对动脉粥样硬化巨噬细胞泡沫化的影响[J].中国中西医结合杂志, 2023, 43(6): 705-711. |
| CHEN X N, GE Q H, ZHAO Y X, et al.Effect of Simiao Yongan Decoction on macrophage foam cell formation in atherosclerosis[J].Chinese Journal of Integrated Traditional and Western Medicine, 2023, 43(6): 705-711. | |
| 17 | XU H, NI Y Q, LIU Y S. Mechanisms of action of miRNAs and lncRNAs in extracellular vesicle in atherosclerosis[J]. Front Cardiovasc Med, 2021, 8: 733985. |
| 18 | RAMILOWSKI J A, GOLDBERG T, HARSHBARGER J, et al. A draft network of ligand-receptor-mediated multicellular signalling in human[J]. Nat Commun, 2015, 6: 7866. |
| 19 | EFREMOVA M, VENTO-TORMO M, TEICHMANN S A, et al. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes[J]. Nat Protoc, 2020, 15(4): 1484-1506. |
| 20 | SHI X, GUO L W, SEEDIAL S M, et al. TGF-β/Smad3 inhibit vascular smooth muscle cell apoptosis through an autocrine signaling mechanism involving VEGF-A[J]. Cell Death Dis, 2014, 5(7): e1317. |
| 21 | OSTRIKER A, HORITA H N, POCZOBUTT J, et al. Vascular smooth muscle cell-derived transforming growth factor-β promotes maturation of activated, neointima lesion-like macrophages[J]. Arterioscler Thromb Vasc Biol, 2014, 34(4): 877-886. |
| 22 | BADIMON L, STOREY R F, VILAHUR G. Update on lipids, inflammation and atherothrombosis[J]. Thromb Haemost, 2011, 105(Suppl 1): S34-S42. |
| 23 | MERCHED A, TOLLEFSON K, CHAN L. β2 integrins modulate the initiation and progression of atherosclerosis in low-density lipoprotein receptor knockout mice[J]. Cardiovasc Res, 2010, 85(4): 853-863. |
| 24 | KANG S W, KIM M S, KIM H S, et al. Celastrol attenuates adipokine resistin-associated matrix interaction and migration of vascular smooth muscle cells[J]. J Cell Biochem, 2013, 114(2): 398-408. |
| 25 | AL BARASHDI M A, ALI A, MCMULLIN M F, et al. Protein tyrosine phosphatase receptor type C (PTPRC or CD45)[J]. J Clin Pathol, 2021, 74(9): 548-552. |
| 26 | 王玉, 孙晓宇, 罗亚, 等. 冠状动脉粥样斑块内CD45表达水平与病灶结构变化的关系[J]. 中国动脉硬化杂志, 2019, 27(2): 114-119, 140. |
| WANG Y, SUN X Y, LUO Y, et al. Relationship between CD45 expression level and lesion structure in coronary artery plaque[J]. Chinese Journal of Arteriosclerosis, 2019, 27(2): 114-119, 140. | |
| 27 | 胡永涛, 刘洪智. 冠心病患者外周血CD45、HMGB1水平与支架内再狭窄的相关性分析[J]. 中国循证心血管医学杂志, 2022, 14(5): 581-584. |
| HU Y T, LIU H Z. Correlation analysis of peripheral blood CD45, HMGB1 and stent restenosis in patients with coronary heart disease[J]. Chinese Journal of Evidence-Based Cardiovascular Medicine, 2022, 14(5): 581-584. | |
| 28 | ZHU S, LIU M, BENNETT S, et al. The molecular structure and role of CCL2 (MCP-1) and C-C chemokine receptor CCR2 in skeletal biology and diseases[J]. J Cell Physiol, 2021, 236(10): 7211-7222. |
| 29 | OSONOI Y, MITA T, AZUMA K, et al. Defective autophagy in vascular smooth muscle cells enhances cell death and atherosclerosis[J]. Autophagy, 2018, 14(11): 1991-2006. |
| 30 | YU B, WONG M M, POTTER C M, et al. Vascular stem/progenitor cell migration induced by smooth muscle cell-derived chemokine (C-C motif) ligand 2 and chemokine (C-X-C motif) ligand 1 contributes to neointima formation[J]. Stem Cells, 2016, 34(9): 2368-2380. |
| 31 | SCHOBER A, ZERNECKE A, LIEHN E A, et al. Crucial role of the CCL2/CCR2 axis in neointimal hyperplasia after arterial injury in hyperlipidemic mice involves early monocyte recruitment and CCL2 presentation on platelets[J]. Circ Res, 2004, 95(11): 1125-1133. |
| 32 | KUNNAS T, SOLAKIVI T, MÄÄTTÄ K, et al. Decorin genotypes, serum glucose, heart rate, and cerebrovascular events: the Tampere adult population cardiovascular risk study[J]. Genet Test Mol Biomarkers, 2016, 20(8): 416-419. |
| 33 | AL HAJ ZEN A, CALIGIURI G, SAINZ J, et al. Decorin overexpression reduces atherosclerosis development in apolipoprotein E-deficient mice[J]. Atherosclerosis, 2006, 187(1): 31-39. |
| 34 | BURTON D G A, GILES P J, SHEERIN A N P, et al. Microarray analysis of senescent vascular smooth muscle cells: a link to atherosclerosis and vascular calcification[J]. Exp Gerontol, 2009, 44(10): 659-665. |
| 35 | FIERRO-MACÍAS A E, FLORIANO-SÁNCHEZ E, MENA-BURCIAGA V M, et al. Association between IGF system and PAPP-A in coronary atherosclerosis[J]. Arch Cardiol Mex, 2016, 86(2): 148-156. |
| 36 | CHONG H, WEI Z, NA M, et al. The PGC-1α/NRF1/miR-378a axis protects vascular smooth muscle cells from FFA-induced proliferation, migration and inflammation in atherosclerosis[J]. Atherosclerosis, 2020, 297: 136-145. |
| 37 | 涂少文, 陈云宪, 唐良秋. 趋化因子在动脉粥样硬化中的作用及研究进展[J]. 中国医学创新, 2022, 19(15): 175-179. |
| TU S W, CHEN Y X, TANG L Q. The role and research progress of chemokines in atherosclerosis[J]. Chinese Medical Innovation, 2022, 19(15): 175-179. | |
| 38 | HERNÁNDEZ-AGUILERA A, FIBLA M, CABRÉ N, et al. Chemokine (C-C motif) ligand 2 and coronary artery disease: tissue expression of functional and atypical receptors[J]. Cytokine, 2020, 126: 154923. |
| 39 | SINGH S R, SUTCLIFFE A, KAUR D, et al. CCL2 release by airway smooth muscle is increased in asthma and promotes fibrocyte migration[J]. Allergy, 2014, 69(9): 1189-1197. |
| 40 | GIRBL T, LENN T, PEREZ L, et al. Distinct compartmentalization of the chemokines CXCL1 and CXCL2 and the atypical receptor ACKR1 determine discrete stages of neutrophil diapedesis[J]. Immunity, 2018, 49(6): 1062-1076.e6. |
| 41 | LIAO Z, JIN Y, CHU Y, et al. Single-cell transcriptome analysis reveals aberrant stromal cells and heterogeneous endothelial cells in alcohol-induced osteonecrosis of the femoral head[J]. Commun Biol, 2022, 5(1): 324. |
| 42 | SPEIDL W S, KASTL S P, HUBER K, et al. Complement in atherosclerosis: friend or foe?[J]. J Thromb Haemost, 2011, 9(3): 428-440. |
| 43 | 刘艾婷, 彭旷, 欧蕾宇, 等. 补体系统在动脉粥样硬化中的作用研究进展[J]. 中国动脉硬化杂志, 2021, 29(4): 363-368. |
| LIU A T, PENG K, OU L Y, et al. Research progress on the role of complement system in atherosclerosis[J]. Chinese Journal of Arteriosclerosis, 2021, 29(4): 363-368. | |
| 44 | WAN J X, FUKUDA N, ENDO M, et al. Complement 3 is involved in changing the phenotype of human glomerular mesangial cells[J]. J Cell Physiol, 2007, 213(2): 495-501. |
| 45 | RUS H, CUDRICI C, NICULESCU F. The role of the complement system in innate immunity[J]. Immunol Res, 2005, 33(2): 103-112. |
| 46 | BUYANNEMEKH D, NHAM S U. Characterization of αX Ⅰ-domain binding to receptors for advanced glycation end products (RAGE)[J]. Mol Cells, 2017, 40(5): 355-362. |
| 47 | YAKUBENKO V P, BHATTACHARJEE A, PLUSKOTA E, et al. αMβ2 integrin activation prevents alternative activation of human and murine macrophages and impedes foam cell formation[J]. Circ Res, 2011, 108(5): 544-554. |
| 48 | BAJTAY Z. Biologia futura: stories about the functions of β2-integrins in human phagocytes[J]. Biol Futur, 2021, 72(1): 7-13. |
| 49 | WANG M, GU M, LIU L, et al. Single-cell RNA sequencing (scRNA-seq) in cardiac tissue: applications and limitations[J]. Vasc Health Risk Manag, 2021, 17: 641-657. |
| [1] | 黄紫晗, 黄心智. 单细胞RNA测序在骨再生研究中的应用[J]. 上海交通大学学报(医学版), 2025, 45(8): 1053-1058. |
| [2] | 朱晗懿, 石欢, 俞创奇, 郑凌艳. 白介素-1β在极重症口腔颌面部感染中的预警价值及机制初探[J]. 上海交通大学学报(医学版), 2025, 45(6): 661-672. |
| [3] | 张星语, 李若谷. 主动脉瘤单细胞转录组的系统性分析与探索[J]. 上海交通大学学报(医学版), 2025, 45(6): 735-744. |
| [4] | 谭颖超, 杨珺玥, 王莉娜. 白细胞介素-1B-511C/T基因多态性与冠状动脉粥样硬化性心脏病关联的meta分析[J]. 上海交通大学学报(医学版), 2022, 42(2): 197-204. |
| [5] | 袁咏梅, 程晓丹, 孙家安, 常东歌, 何莹莹, 刘畅. lncRNA NEAT1通过miR-377-3p/Wnt通路影响ox-LDL诱导的人血管平滑肌细胞增殖、侵袭迁移[J]. 上海交通大学学报(医学版), 2022, 42(11): 1534-1541. |
| [6] | 刘霞, 温弗乐, 章雅青. 冠心病患者参加门诊心脏康复的障碍水平调研及相关因素分析[J]. 上海交通大学学报(医学版), 2022, 42(10): 1448-1457. |
| [7] | 邢海帆, 范瑛. 单细胞RNA测序应用于肾小球疾病研究的进展[J]. 上海交通大学学报(医学版), 2022, 42(10): 1458-1465. |
| [8] | 李雨晨, 白丽莲, 黄荷凤, 刘欣梅. 人胸腺基质发育及退化的单细胞转录组分析[J]. 上海交通大学学报(医学版), 2021, 41(7): 865-875. |
| [9] | 罗嘉强, 赵亮宇, 姚晨成, 朱子珏, 邢晓宇, 李朋, 田汝辉, 陈慧兴, 孙杰, 李铮. 基于单细胞RNA测序解析人与小鼠睾丸中SARS-CoV-2相关受体的时空表达特征[J]. 上海交通大学学报(医学版), 2021, 41(4): 421-426. |
| [10] | 赵倩, 高霖, 王长谦, 张俊峰, 张绘莉, 卓杨. 冠状动脉粥样硬化性心脏病患者外周血单核细胞亚群CX3CR1表达的变化及意义[J]. 上海交通大学学报(医学版), 2021, 41(3): 328-333. |
| [11] | 马燕如, 季林华, 童天颖, 严宇青, 沈超琴, 张昕雨, 曹颖颖, 洪洁, 陈豪燕. 基于单细胞RNA测序的结直肠癌预后预测模型的建立和验证[J]. 上海交通大学学报(医学版), 2021, 41(2): 159-165. |
| [12] | 范益博1, 2,幺天保1,马 珺1,袁安彩1,杜勇平1,邵 琴1,卜 军1. 血清维生素D与冠状动脉粥样硬化性心脏病患者冠状动脉病变严重程度及短期预后的相关性研究[J]. 上海交通大学学报(医学版), 2020, 40(7): 889-893. |
| [13] | 路芳芳1,姚佳璐2,钮晓音3,翁 震4#,何 杨1, 4#. 阿苯达唑对小鼠动脉血管狭窄的干预效果及机制研究[J]. 上海交通大学学报(医学版), 2020, 40(5): 598-603. |
| [14] | 徐 洪*,朱鹏雄*,周衍再,裘佳培,刘 俊#,赵 强#. 微创冠状动脉旁路移植术的远期疗效分析[J]. 上海交通大学学报(医学版), 2020, 40(5): 656-661. |
| [15] | 李 鑫*,梁馨月*,方宁远,汪海娅. 老年稳定性冠状动脉粥样硬化性心脏病患者合并衰弱综合征的影响因素分析[J]. 上海交通大学学报(医学版), 2020, 40(12): 1627-1631. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||