
上海交通大学学报(医学版) ›› 2024, Vol. 44 ›› Issue (9): 1124-1135.doi: 10.3969/j.issn.1674-8115.2024.09.007
收稿日期:2024-04-28
接受日期:2024-07-16
出版日期:2024-09-28
发布日期:2024-09-28
通讯作者:
聂惠贞,电子信箱:hznie@shsci.org。作者简介:钱立恒(1998—),男,硕士生;电子信箱:shandianqianliheng@sjtu.edu.cn。
基金资助:
QIAN Liheng( ), WEN Kailing, LIAO Yingna, LI Shuxin, NIE Huizhen(
), WEN Kailing, LIAO Yingna, LI Shuxin, NIE Huizhen( )
)
Received:2024-04-28
Accepted:2024-07-16
Online:2024-09-28
Published:2024-09-28
Contact:
NIE Huizhen, E-mail: hznie@shsci.org.Supported by:摘要:
目的·分析分选链接蛋白1(sorting nexin 1,SNX1)在结直肠癌(colorectal cancer,CRC)中的表达,探索其对CRC细胞增殖及迁移的影响及潜在的分子机制。方法·基于癌症基因组图谱(The Cancer Genome Atlas,TCGA)、基因型-组织表达(Genotype-Tissue Expression,GTEx)以及基因表达综合(Gene Expression Omnibus,GEO)数据库中CRC相关的转录组数据和临床病理信息,利用基因集富集分析(Gene Set Enrichment Analysis,GSEA)软件进行富集分析。采用实时荧光定量聚合酶链反应(quantitative real-time polymerase chain reaction,qPCR)、蛋白质印迹法(Western blotting)、免疫组织化学染色(immunohistochemistry staining,IHC)检测SNX1在CRC组织和细胞中的表达。使用小干扰RNA(small interfering RNA,siRNA)敲低SNX1的表达,观察SNX1对肿瘤细胞增殖和迁移能力的影响。通过相关性分析探索SNX1影响CRC细胞迁移的潜在分子机制,在SNX1敲低细胞株中进行mRNA水平的初步验证。结果·根据对TCGA中CRC患者以及组织芯片样本数据的分析,发现SNX1在CRC组织中表达下调,且与肿瘤的直径以及是否发生远处转移具有相关性。敲低SNX1能够促进肿瘤细胞的增殖和迁移。在CRC中,SNX1的表达与结肠癌转移相关因子1(metastasis associated in colon cancer 1,MACC1)、间质-上皮转换因子(mesenchymal to epithelial transition factor,MET)以及Notch负相关,敲低SNX1后上述基因表达上调;敲低SNX1后,上皮-间质转化(epithelial to mesenchymal transition,EMT)标志物钙黏蛋白1(cadherin 1,CDH1)表达下调,波形蛋白(vimentin,VIM)、Snail家族转录因子1(Snail family transcriptional repressor 1,SNAI1)表达上调。结论·SNX1在CRC组织中表达显著降低,且与患者预后正相关;SNX1低表达能够促进CRC细胞的增殖和迁移,与MACC1-MET通路、EMT相关;SNX1可作为CRC不良预后的潜在生物标志物和新的治疗靶点。
中图分类号:
钱立恒, 温凯玲, 廖颖娜, 李书鑫, 聂惠贞. 分选链接蛋白1抑制结直肠癌细胞增殖和迁移的作用和机制研究[J]. 上海交通大学学报(医学版), 2024, 44(9): 1124-1135.
QIAN Liheng, WEN Kailing, LIAO Yingna, LI Shuxin, NIE Huizhen. Study on the effect and mechanism of sorting nexin 1 on inhibiting the proliferation and migration of colorectal cancer cells[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2024, 44(9): 1124-1135.
| siRNA | Forward (5′➝3′) | Reverse (5′➝3′) |
|---|---|---|
| NC-siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
| SNX1-siRNA1 | GAGGAUCAAUUUGAUUUGATT | UCAAAUCAAAUUGAUCCUCTT |
| SNX1-siRNA2 | CAGGCCAACAAUGACUUCUTT | AGAAGUCAUUGUUGGCCUGTT |
表1 siRNA序列
Tab 1 Sequences of siRNA
| siRNA | Forward (5′➝3′) | Reverse (5′➝3′) |
|---|---|---|
| NC-siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
| SNX1-siRNA1 | GAGGAUCAAUUUGAUUUGATT | UCAAAUCAAAUUGAUCCUCTT |
| SNX1-siRNA2 | CAGGCCAACAAUGACUUCUTT | AGAAGUCAUUGUUGGCCUGTT |
| Primer | Forward (5′➝3′) | Reverse (5′➝3′) |
|---|---|---|
| 18s mRNA | TGCGAGTACTCAACACCAACA | GCATATCTTCGGCCCACA |
| SNX1 | AAGCACTCTCAGAATGGCTTC | CGGCCCTCCGTTTTTCAAG |
| MACC1 | GGGTCACAGGTGAACGAGAT | CTGGGTCCTGGCATTCTGTA |
| MET | ATTTTGCTTTGCCAGTGGTGG | AGCGATGTTGACATGCCACT |
| NOTCH1 | AGAGGCGTGGCAGACTATG | CTGGCACGATTTCCCTGACC |
| CDH1 | GGCTGGACCGAGAGAGTTTC | CGACGTTAGCCTCGTTCTCA |
| VIM | CTCCCTGAACCTGAGGGAAAC | TTGCGCTCCTGAAAAACTGC |
| SNAI1 | TAGCGAGTGGTTCTTCTGCG | TGCTGGAAGGTAAACTCTGGA |
表2 qPCR引物序列
Tab 2 Primer Sequences for qPCR
| Primer | Forward (5′➝3′) | Reverse (5′➝3′) |
|---|---|---|
| 18s mRNA | TGCGAGTACTCAACACCAACA | GCATATCTTCGGCCCACA |
| SNX1 | AAGCACTCTCAGAATGGCTTC | CGGCCCTCCGTTTTTCAAG |
| MACC1 | GGGTCACAGGTGAACGAGAT | CTGGGTCCTGGCATTCTGTA |
| MET | ATTTTGCTTTGCCAGTGGTGG | AGCGATGTTGACATGCCACT |
| NOTCH1 | AGAGGCGTGGCAGACTATG | CTGGCACGATTTCCCTGACC |
| CDH1 | GGCTGGACCGAGAGAGTTTC | CGACGTTAGCCTCGTTCTCA |
| VIM | CTCCCTGAACCTGAGGGAAAC | TTGCGCTCCTGAAAAACTGC |
| SNAI1 | TAGCGAGTGGTTCTTCTGCG | TGCTGGAAGGTAAACTCTGGA |
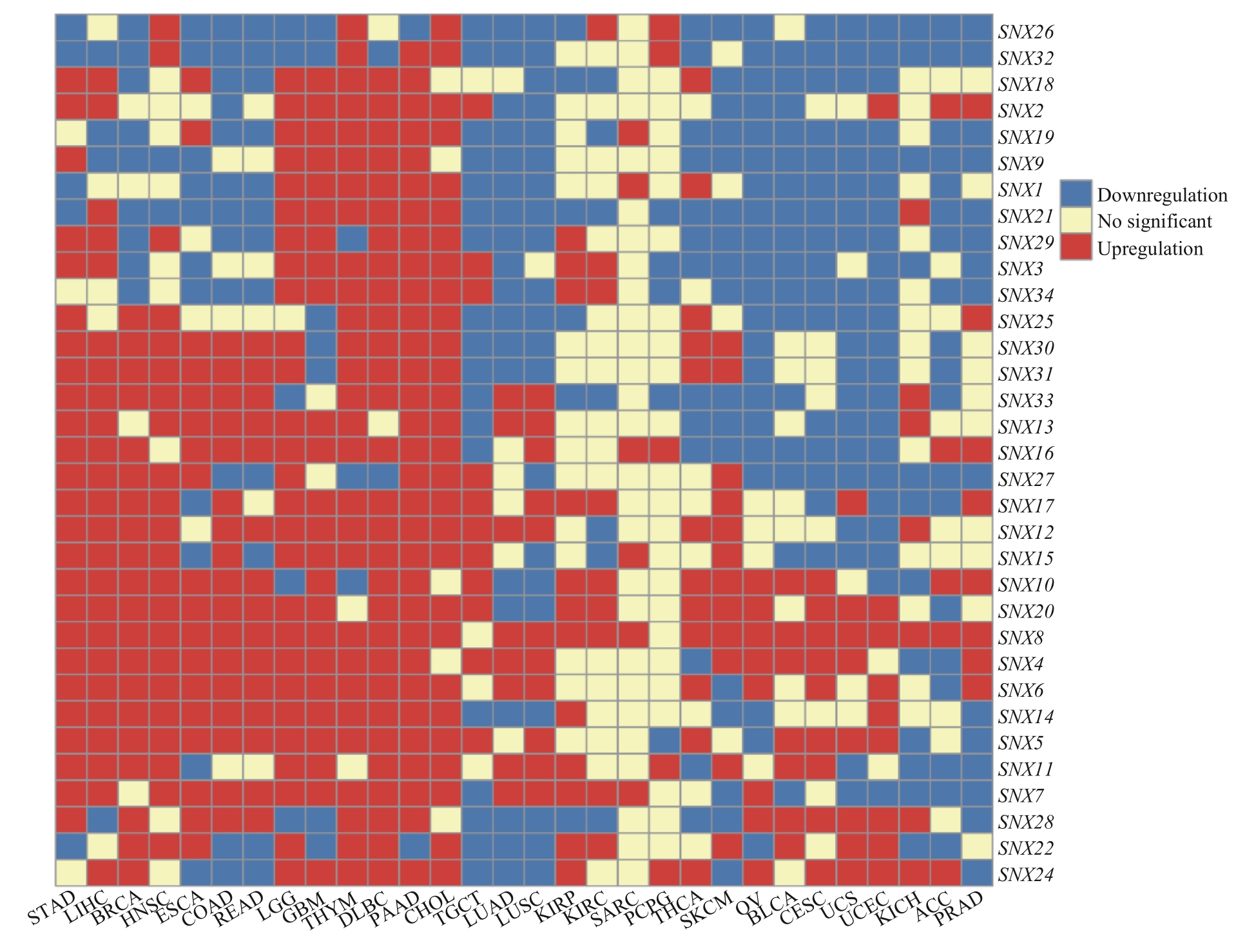
图1 SNX家族在人类肿瘤中的表达情况Note: STAD—stomach adenocarcinoma; LIHC—liver hepatocellular carcinoma; BRCA—breast invasive carcinoma; HNSC—head and neck squamous cell carcinoma; ESCA—esophageal carcinoma; COAD—colon adenocarcinoma; READ—rectum adenocarcinoma; LGG—brain lower grade glioma; GBM—glioblastoma multiforme; THYM—thymoma; DLBC—lymphoid neoplasm diffuse large B-cell lymphoma; PAAD—pancreatic adenocarcinoma; CHOL—cholangiocarcinoma; TGCT—testicular germ cell tumor; LUAD—lung adenocarcinoma; LUSC—lung squamous cell carcinoma; KIRP—kidney renal papillary cell carcinoma; KIRC—kidney renal clear cell carcinoma; SARC—sarcoma; PCPG—pheochromocytoma and paraganglioma; THCA—thyroid carcinoma; SKCM—skin cutaneous melanoma; OV—ovarian serous cystadenocarcinoma; BLCA—bladder urothelial carcinoma; CESC—cervical squamous cell carcinoma and endocervical adenocarcinoma; UCS—uterine carcinosarcoma; UCEC—uterine corpus endometrial carcinoma; KICH—kidney chromophobe; ACC—adrenocortical carcinoma; PRAD—prostate adenocarcinoma.
Fig 1 Expression levels of SNX family in human cancers
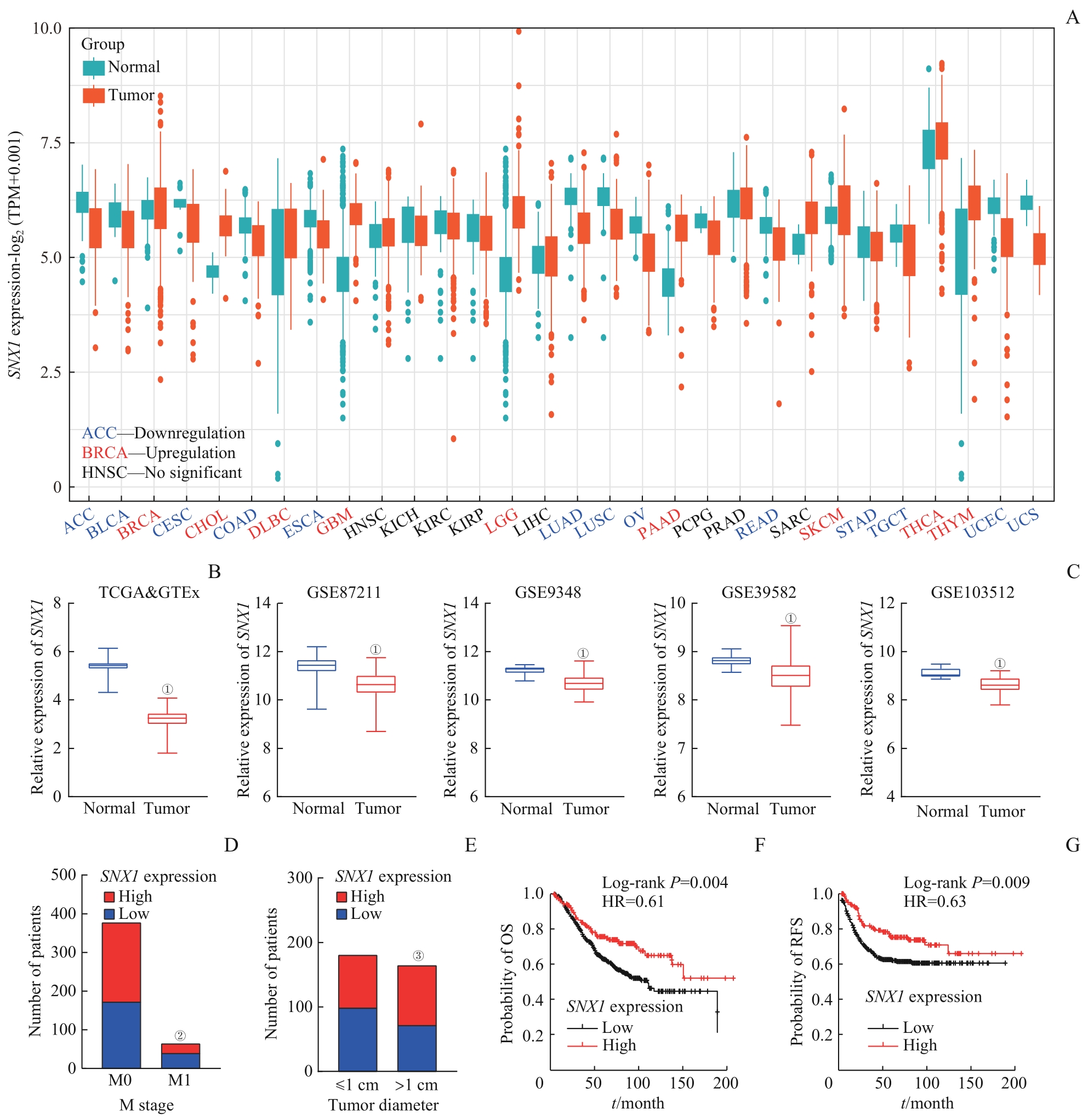
图2 SNX1 在CRC数据库中表达情况及其与临床病理信息的相关性分析Note: A. Expression levels of SNX1 mRNA in human cancers. TPM—transcript per million. B. Expression levels of SNX1 mRNA in CRC tissues and normal colon tissues from the TCGA and GTEx databases. C. Expression levels of SNX1 mRNA in CRC tissues and normal colon tissues from the GEO databases. D. Correlation between SNX1 mRNA expression and M stage of patients from the TCGA database. E. Correlation between SNX1 mRNA expression and tumor size in patients from the TCGA database. F. Correlation between SNX1 mRNA expression and overall survival (OS) in patients from the GSE39582 database. G. Correlation between SNX1 mRNA expression and recurrence-free survival (RFS) in patients from the GSE39582 database. ①P=0.000, compared with normal colon tissues; ②P=0.022, compared with M0 groups; ③P=0.041, compared with the ≤1 cm groups.
Fig 2 Expression levels of SNX1 from the CRC database and its correlation with clinical pathological information
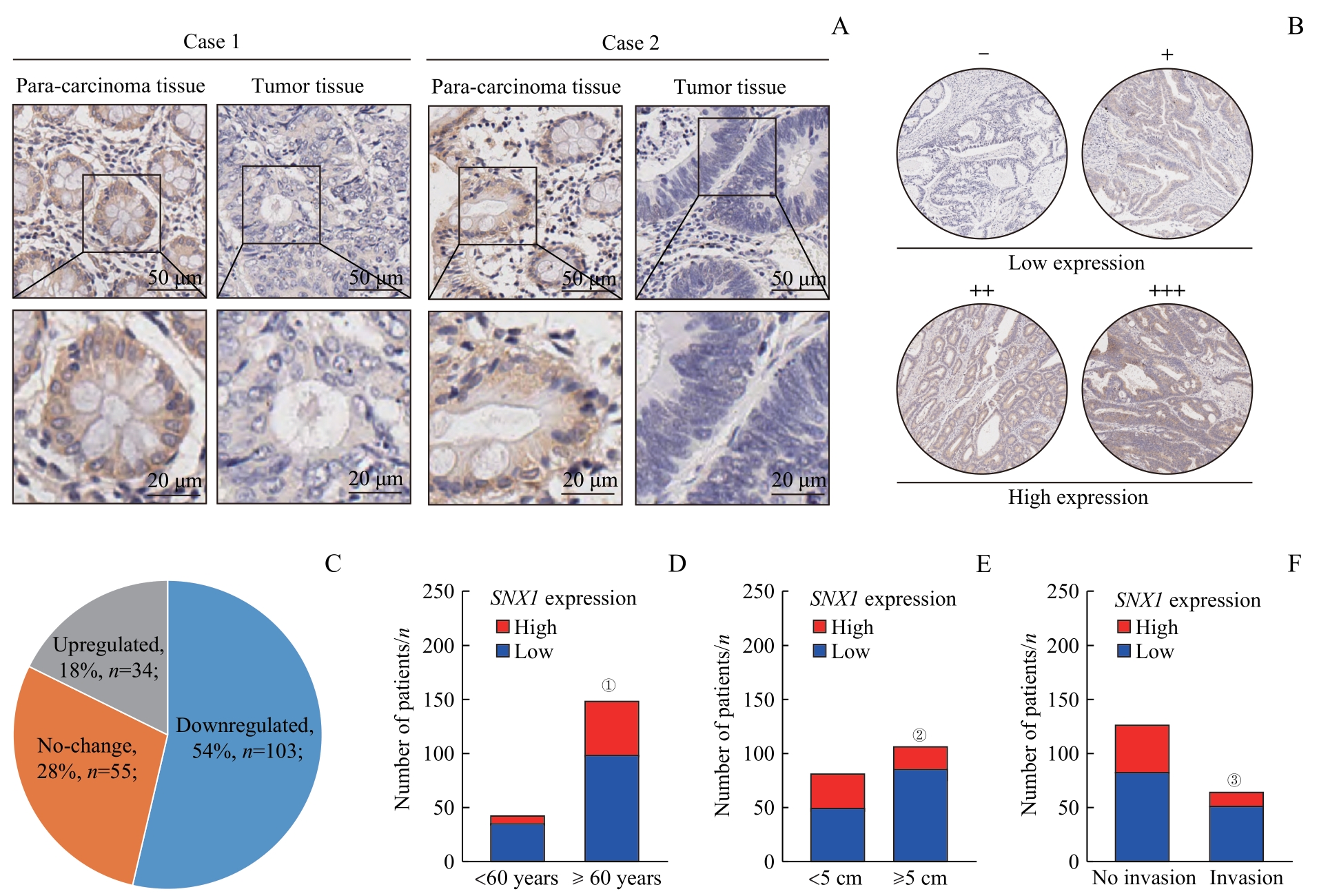
图3 SNX1在CRC组织中表达情况及其与临床病理信息的相关性分析Note: A. Expression of SNX1 detected by IHC in CRC tissues and para-carcinoma tissues from CRC patients. B. SNX1 staining was divided into four grades according to the depth of tissue microarray IHC staining. The two lower grades were grouped as the low-expression group, while the two higher grades were classified as the high-expression group. C. Expression of SNX1 was decreased in 54% of CRC tissues in tissue microarrays, by comparing IHC grades of CRC tissues with para-carcinoma tissues D. Correlation between SNX1 protein expression and age of patients in tissue microarrays. E. Correlation between SNX1 protein expression and tumor size of patients in tissue microarrays. F. Correlation between SNX1 protein expression and lymphatic invasion of patients in tissue microarrays. ①P=0.036, compared with the <60 years old group; ②P=0.005, compared with the <5 cm group; ③P=0.045, compared with the no lymphatic invasion group.
Fig 3 Expression levels of SNX1 in CRC tissues and their correlation with clinical pathological information
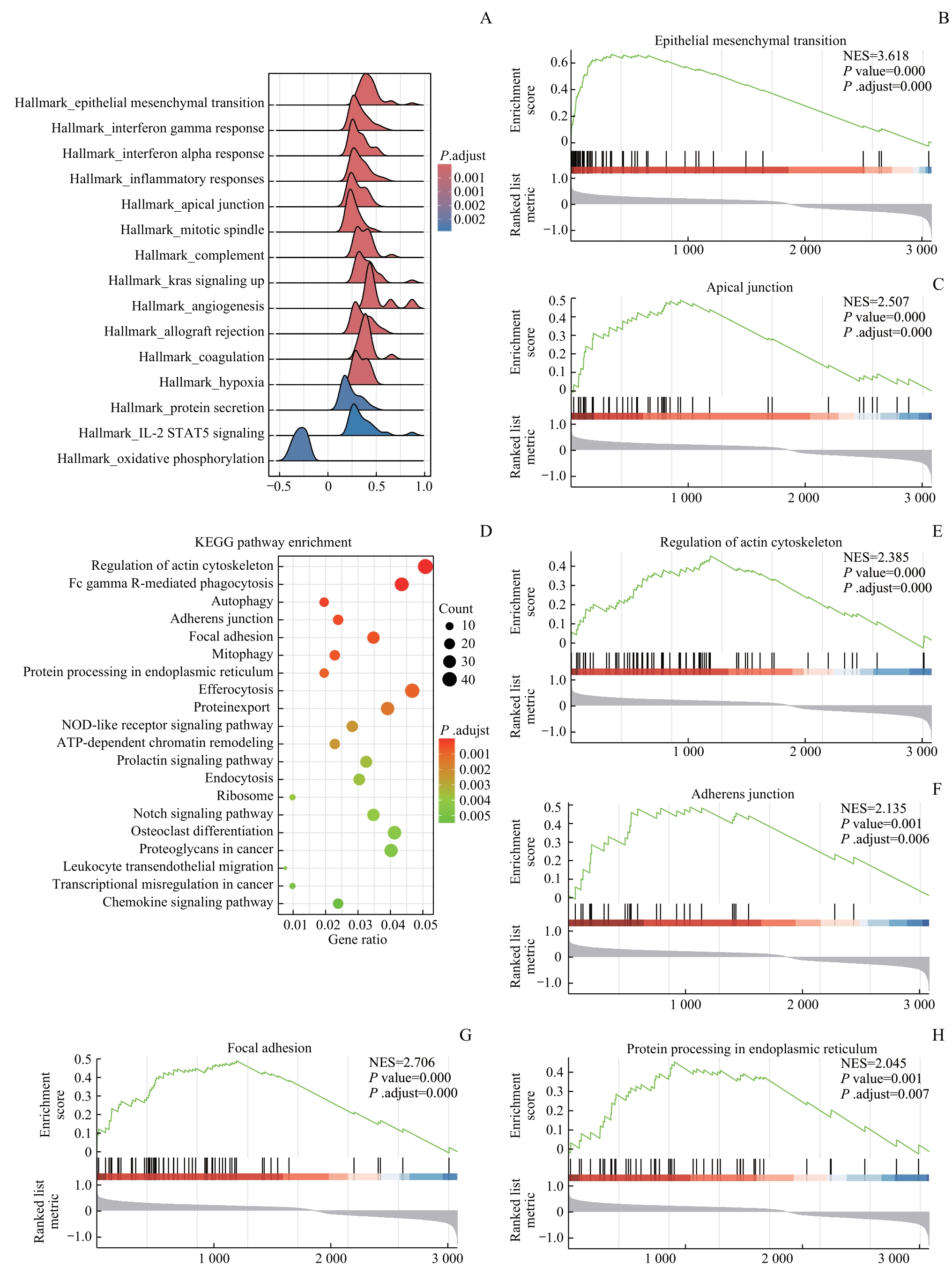
图4 SNX1相关信号通路的富集分析Note: A. Ridgeline plots of Hallmark enrichment analysis based on SNX1 low-expression and high-expression groups. B/C. GSEA showed that EMT signaling pathway (B) and apical junction pathway (C) were enriched in the SNX1 low-expression group. D. Dot plots of KEGG enrichment analysis based on the SNX1 low-expression and high-expression groups. E?H. GSEA showed that regulation of actin cytoskeleton pathway (E), adherens junction pathway (F), focal adhesion (G) and protein processing in endoplasmic reticulum pathway (H) were enriched in the SNX1 low-expression group.
Fig 4 Enrichment analysis of SNX1-related signaling pathways

图5 SNX1的表达水平对CRC细胞增殖和迁移的影响Note: A. Expression levels of SNX1 detected by qPCR and western blotting in CRC cell lines. B/C. Transfection efficiency of SNX1-siRNA verified by qPCR (top) and western blotting (bottom) in HT29 cells (B) and SW480 cells (C). D/E. Effect of SNX1-siRNA knockdown on HT29 cell (D) and SW480 cell (E) proliferation detected by CCK-8. F/G. Effect of SNX1-siRNA knockdown on HT29 cell (F) and SW480 cell (G) migration (left) detected by wound healing assay and analysis of migration rate (right). H. Effect of SNX1-siRNA knockdown on HT29 cell (top) and SW480 cell (bottom) migration (left) detected by Transwell assay and analysis of migration cells (right). I. Overexpression efficiency of SNX1-OE verified by western blotting in LoVo cells. J. Effect of SNX1-OE overexpression on LoVo cell proliferation detected by CCK-8. K. Effect of SNX1-OE overexpression on LoVo cell migration (left) detected by wound healing assay and analysis of migration rate (right). L. Effect of SNX1-OE overexpression on LoVo cell migration (left) detected by Transwell assay and analysis of migration cells (right). ①P=0.000, ②P=0.003, ③P=0.000, ④P=0.005, compared with the NC-siRNA group; ⑤P=0.000, ⑥P=0.003, compared with the SNX1-vector group.
Fig 5 Effect of SNX1 expression on the proliferation and migration of CRC cells
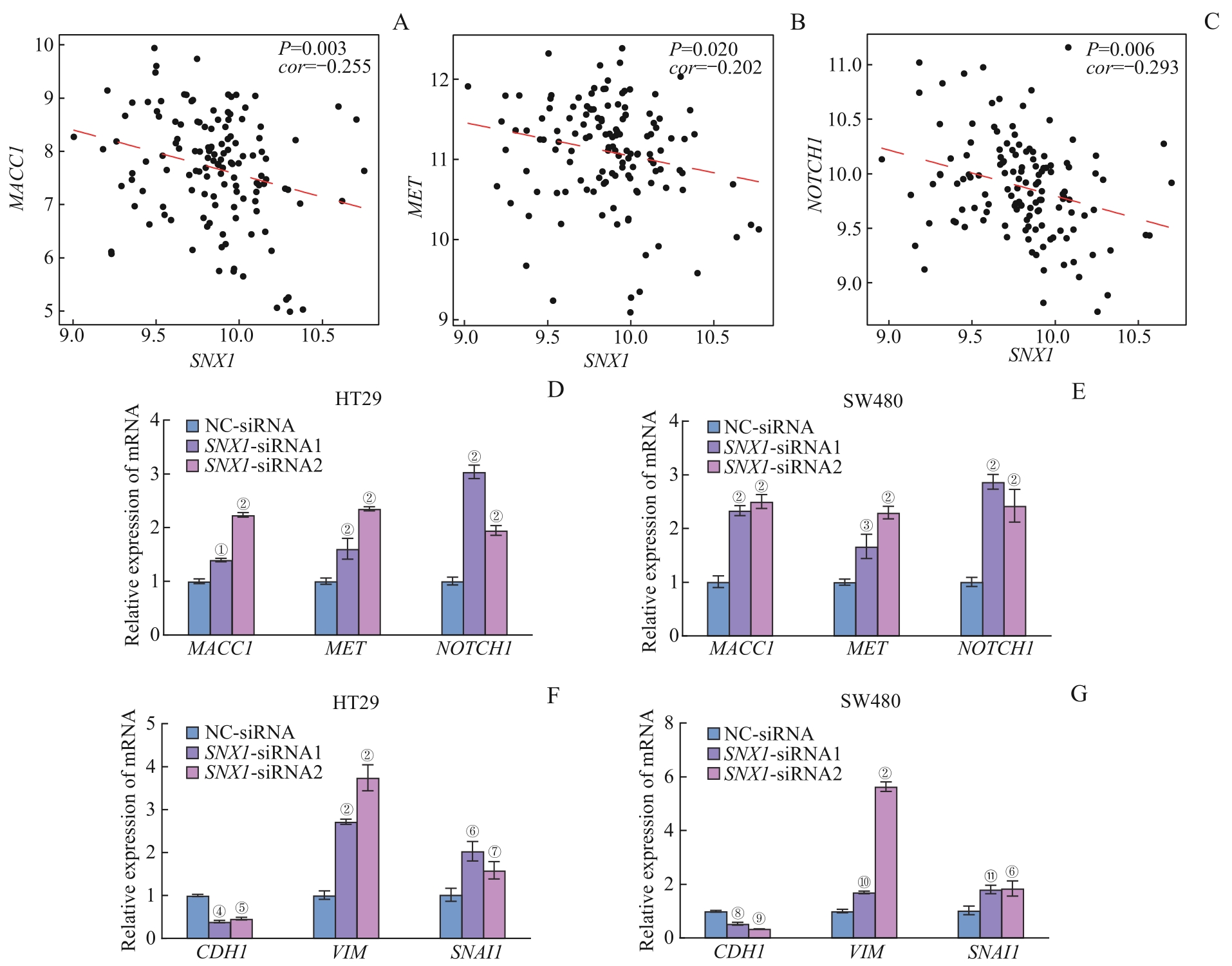
图6 SNX1 低表达与CRC转移关键基因以及EMT相关Note: A?C. Correlation analysis between SNX1 and MACC1 (A), MET (B), NOTCH1 (C) in GSE41568 database. D/E. Relative mRNA expression of MACC1, MET and NOCTH1 in SNX1-konckdown and control groups in HT29 cells (D) and SW480 cells (E). F/G. Relative mRNA expression of CDH1, VIM and SNAI1 in SNX1-konckdown and control groups in HT29 cells (F) and SW480 cells (G). ①P=0.013, ②P=0.000, ③P=0.015, ④P=0.025, ⑤P=0.048, ⑥P=0.001, ⑦P=0.037, ⑧P=0.047, ⑨P=0.006, ⑩P=0.004, ?P=0.002, compared with the NC-siRNA group.
Fig 6 Correlation between low expression of SNX1 and key genes of CRC metastasis and EMT
| 1 | WADHWA V, PATEL N, GROVER D, et al. Interventional gastroenterology in oncology[J]. CA Cancer J Clin, 2023, 73(3): 286-319. |
| 2 | SIEGEL R L, MILLER K D, GODING SAUER A, et al. Colorectal cancer statistics, 2020[J]. CA Cancer J Clin, 2020, 70(3): 145-164. |
| 3 | VAN CUTSEM E, NORDLINGER B, ADAM R, et al. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases[J]. Eur J Cancer, 2006, 42(14): 2212-2221. |
| 4 | QI Z, LAZAR C S, TRONCHÈRE H, et al. Endosomal localization and function of sorting nexin 1[J]. Proc Natl Acad Sci USA, 2002, 99(10): 6767-6772. |
| 5 | KURTEN R C, CADENA D L, GILL G N. Enhanced degradation of EGF receptors by a sorting nexin, SNX1[J]. Science, 1996, 272(5264): 1008-1010. |
| 6 | NISHIMURA Y, YOSHIOKA K, BERECZKY B, et al. Evidence for efficient phosphorylation of EGFR and rapid endocytosis of phosphorylated EGFR via the early/late endocytic pathway in a gefitinib-sensitive non-small cell lung cancer cell line[J]. Mol Cancer, 2008, 7: 42. |
| 7 | ZHAN X Y, ZHANG Y Q, ZHAI E T, et al. Sorting nexin-1 is a candidate tumor suppressor and potential prognostic marker in gastric cancer[J]. PeerJ, 2018, 6: e4829. |
| 8 | 潘泓, 廖颖娜, 盖严支, 等. 分选链接蛋白1在胰腺导管腺癌中的表达及其促进胰腺导管腺癌进展的机制研究[J]. 上海交通大学学报(医学版), 2023, 43(3): 278-292. |
| PAN H, LIAO Y N, GAI Y Z, et al. Expression of sorting nexin 1 in pancreatic ductal adenocarcinoma and its mechanism in promoting PDAC progress[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2023, 43(3): 278-292. | |
| 9 | HUANG Y H, HONG W Q, WEI X W. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis[J]. J Hematol Oncol, 2022, 15(1): 129. |
| 10 | BILLER L H, SCHRAG D. Diagnosis and treatment of metastatic colorectal cancer: a review[J]. JAMA, 2021, 325(7): 669-685. |
| 11 | KAHI C J, BOLAND C R, DOMINITZ J A, et al. Colonoscopy surveillance after colorectal cancer resection: recommendations of the US multi-society task force on colorectal cancer[J]. Am J Gastroenterol, 2016, 111(3): 337-346; quiz 347. |
| 12 | GERSTBERGER S, JIANG Q W, GANESH K. Metastasis[J]. Cell, 2023, 186(8): 1564-1579. |
| 13 | ZHAO Y, LIU Y J, LIN L, et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1[J]. Mol Cancer, 2018, 17(1): 69. |
| 14 | RADHAKRISHNAN H, WALTHER W, ZINCKE F, et al. MACC1: the first decade of a key metastasis molecule from gene discovery to clinical translation[J]. Cancer Metastasis Rev, 2018, 37(4): 805-820. |
| 15 | NISHIMURA Y, TAKIGUCHI S, ITO S, et al. Evidence that depletion of the sorting nexin 1 by siRNA promotes HGF-induced MET endocytosis and MET phosphorylation in a gefitinib-resistant human lung cancer cell line[J]. Int J Oncol, 2014, 44(2): 412-426. |
| 16 | YUAN X, WU H, HAN N, et al. Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application[J]. J Hematol Oncol, 2014, 7: 87. |
| 17 | WANG Y D, WU B R, FARRAR E, et al. Notch-Tnf signalling is required for development and homeostasis of arterial valves[J]. Eur Heart J, 2017, 38(9): 675-686. |
| 18 | KIM R K, KAUSHIK N, SUH Y, et al. Radiation driven epithelial-mesenchymal transition is mediated by Notch signaling in breast cancer[J]. Oncotarget, 2016, 7(33): 53430-53442. |
| 19 | ZAVADIL J, CERMAK L, SOTO-NIEVES N, et al. Integration of TGF-β/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition[J]. EMBO J, 2004, 23(5): 1155-1165. |
| [1] | 朱子俊, 钱逸斐, 李倩玉, 李松玲, 覃雯莉, 刘艳丰. 后期促进复合体亚基10调控PI3K-AKT-mTOR通路促进肝细胞癌进展的研究[J]. 上海交通大学学报(医学版), 2025, 45(9): 1171-1182. |
| [2] | 江怡, 黄晨浩, 李祉良, 吴珺玮, 赵任, 张弢. 1例KRAS突变的结直肠癌患者术前接受化疗联合免疫治疗的效果报道[J]. 上海交通大学学报(医学版), 2025, 45(9): 1256-1260. |
| [3] | 许沐馨, 刘贤, 蒋立姗, 孙青. Nd:YAP激光生物刺激通过WNT/β-catenin信号通路促进人牙周韧带细胞增殖和成骨分化[J]. 上海交通大学学报(医学版), 2025, 45(5): 562-569. |
| [4] | 陈怡楠, 郑旸, 曾汉林, 雷鸣. Fas相关死亡结构域蛋白促进头颈部鳞状细胞癌细胞增殖能力的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(4): 404-414. |
| [5] | 陈佳莹, 褚以忞, 彭海霞. 结直肠癌无进展生存时间预测模型及影响因素研究[J]. 上海交通大学学报(医学版), 2025, 45(3): 324-334. |
| [6] | 梁乐斌, 陈慧芳, 赖淑静, 顾靓, 苏冰. 基于空间ATAC-seq技术的Apcmin/+小鼠结肠肿瘤表观特征分析[J]. 上海交通大学学报(医学版), 2025, 45(10): 1261-1270. |
| [7] | 张先洲, 杜凤麟, 吴雷, 任逸喆, 赵明娜, 娄加陶. OGT通过ERK信号通路促进非小细胞肺癌增殖的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(10): 1288-1297. |
| [8] | 李想, 魏鸣, 吴文曦, 罗小琴, 姚彪, 伍思宇. 芦丁对骨肉瘤生长和转移的体内外抑制作用[J]. 上海交通大学学报(医学版), 2025, 45(1): 20-28. |
| [9] | 孙晨玮, 海汪溪, 屈骞, 席云. [18F]F-FMISO和[18F]F-FLT PET/CT双核素显像预测胰腺癌耐药性的体内研究[J]. 上海交通大学学报(医学版), 2025, 45(1): 60-68. |
| [10] | 施灵玲, 程燕咏, 张磊. 七氟烷对原代少突胶质细胞增殖和分化的影响[J]. 上海交通大学学报(医学版), 2024, 44(9): 1115-1123. |
| [11] | 谭露, 沈少明, 何平. 低氧诱导的长链非编码RNA 68在肝癌中的功能及其作用机制[J]. 上海交通大学学报(医学版), 2024, 44(6): 702-712. |
| [12] | 冯昫皎, 刘健悦, 戚炀炀, 孙晶, 沈蕾. 结直肠癌中自然杀伤细胞表型及功能初探[J]. 上海交通大学学报(医学版), 2024, 44(6): 713-722. |
| [13] | 俞洋, 孟丹, 仇奕文, 袁见, 朱莹杰. 两样本孟德尔随机化法分析1型糖尿病对结直肠癌的影响[J]. 上海交通大学学报(医学版), 2024, 44(6): 755-761. |
| [14] | 蔡人杰, 徐明. KHSRP通过ANK3调节前列腺癌细胞对雄激素的反应性[J]. 上海交通大学学报(医学版), 2024, 44(4): 417-426. |
| [15] | 安俊伊, 陈必颖, 陈循睿, 尹姗姗, 边洲亮, 刘峰. SFXN3在头颈部鳞状细胞癌中的表达及其对细胞增殖的影响[J]. 上海交通大学学报(医学版), 2024, 44(4): 427-434. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||