
上海交通大学学报(医学版) ›› 2021, Vol. 41 ›› Issue (9): 1197-1206.doi: 10.3969/j.issn.1674-8115.2021.09.010
徐莹( ), 褚以忞, 杨大明, 李吉, 张海芹, 彭海霞(
), 褚以忞, 杨大明, 李吉, 张海芹, 彭海霞( )
)
收稿日期:2021-03-10
出版日期:2021-09-28
发布日期:2021-08-24
通讯作者:
彭海霞,电子信箱:phx1101@shtrhospital.com。作者简介:徐莹(1987—),女,主治医师,博士生;电子信箱:xy3459@shtrhospital.com。
基金资助:
Ying XU( ), Yi-min CHU, Da-ming YANG, Ji LI, Hai-qin ZHANG, Hai-xia PENG(
), Yi-min CHU, Da-ming YANG, Ji LI, Hai-qin ZHANG, Hai-xia PENG( )
)
Received:2021-03-10
Online:2021-09-28
Published:2021-08-24
Contact:
PENG Hai-xia, E-mail: phx1101@shtrhospital.com.Supported by:摘要:
目的·从转录组层面探究影响高度微卫星不稳定(microsatellite instability-high,MSI-H)结直肠癌转移的潜在关键基因及基因表达特征,并构建基因转移预测模型。方法·从癌症基因组图谱数据库中收集MSI-H结直肠癌患者转录组数据,根据转移信息分为转移组(21例)和无转移组(42例),分析2组间差异表达基因(differentially expressed gene,DEG),以基因本体数据库(Gene Ontology,GO)、基因集富集分析(Gene Set Enrichment Analysis,GSEA)对DEG进行注释、聚类及信号通路富集;使用STRING、Cytoscape软件筛选枢纽基因(hub基因);选取DEG绘制列线图,使用Bootstrap方法进行交叉验证;分析列线图中每个基因对MSI-H结直肠癌无进展生存期(progression-free survival,PFS)的影响。结果·转移组和无转移组间共得到245个DEGs,其中转移组较无转移组表达上调基因204个,下调基因41个。GO分析发现:DEG在生物过程、分子功能上主要富集于离子穿膜转运、氯离子穿膜转运及氯离子通道活性;在细胞组分中,富集于细胞外部分、细胞外空间等。GSEA结果显示:上调基因富集于神经活性物质配体-受体相互作用和代谢信号通路。通过Cytoscape筛选出上调基因蛋白质互作网络中的前10位的hub基因。根据DEG中调整后P值最小且与肿瘤发生发展关联性高的前10个基因构建的转移预测模型有一定的预测效能,其中训练集曲线下面积(area under curve,AUC)=0.975,验证集AUC=0.920;模型中AC078993.1、IGLJ2(immunoglobulin lambda joining 2)的表达水平与MSI-H结直肠癌PFS呈明显负相关(P=0.011,P=0.005)。结论·在MSI-H结直肠癌中,离子通道变化及细胞外环境变化可能对肿瘤转移有重要影响,神经活性物质配体-受体相互作用、代谢信号通路可能是对转移较重要的信号通路;初步构建了MSI-H结直肠癌基因转移预测模型,可为后续相关临床研究提供参考。
中图分类号:
徐莹, 褚以忞, 杨大明, 李吉, 张海芹, 彭海霞. 基于差异表达基因组合构建高度微卫星不稳定结直肠癌转移预测模型[J]. 上海交通大学学报(医学版), 2021, 41(9): 1197-1206.
Ying XU, Yi-min CHU, Da-ming YANG, Ji LI, Hai-qin ZHANG, Hai-xia PENG. Construction of a metastasis prediction model of microsatellite instability-high colorectal cancer based on differentially expressed gene assembly[J]. JOURNAL OF SHANGHAI JIAOTONG UNIVERSITY (MEDICAL SCIENCE), 2021, 41(9): 1197-1206.
| Characteristic | Metastasis group (n=21) | Non-metastasis group (n=42) |
|---|---|---|
| Gender/n(%) | ||
| Female | 12 (57.1) | 22 (52.4) |
| Male | 9 (42.9) | 20 (47.6) |
| Race/n(%) | ||
| Unknown | 9 (42.9) | 7 (16.7) |
| Asian | 0 (0) | 1 (2.4) |
| Black or African American | 2 (9.5) | 7 (16.7) |
| White | 10 (47.6) | 27 (64.3) |
| Survival status/n(%) | ||
| Alive | 17 (81.0) | 41 (97.6) |
| Dead | 4 (19.0) | 1 ( 2.4) |
表1 63例纳入患者基本信息
Tab 1 Basic information of 63 included patients
| Characteristic | Metastasis group (n=21) | Non-metastasis group (n=42) |
|---|---|---|
| Gender/n(%) | ||
| Female | 12 (57.1) | 22 (52.4) |
| Male | 9 (42.9) | 20 (47.6) |
| Race/n(%) | ||
| Unknown | 9 (42.9) | 7 (16.7) |
| Asian | 0 (0) | 1 (2.4) |
| Black or African American | 2 (9.5) | 7 (16.7) |
| White | 10 (47.6) | 27 (64.3) |
| Survival status/n(%) | ||
| Alive | 17 (81.0) | 41 (97.6) |
| Dead | 4 (19.0) | 1 ( 2.4) |
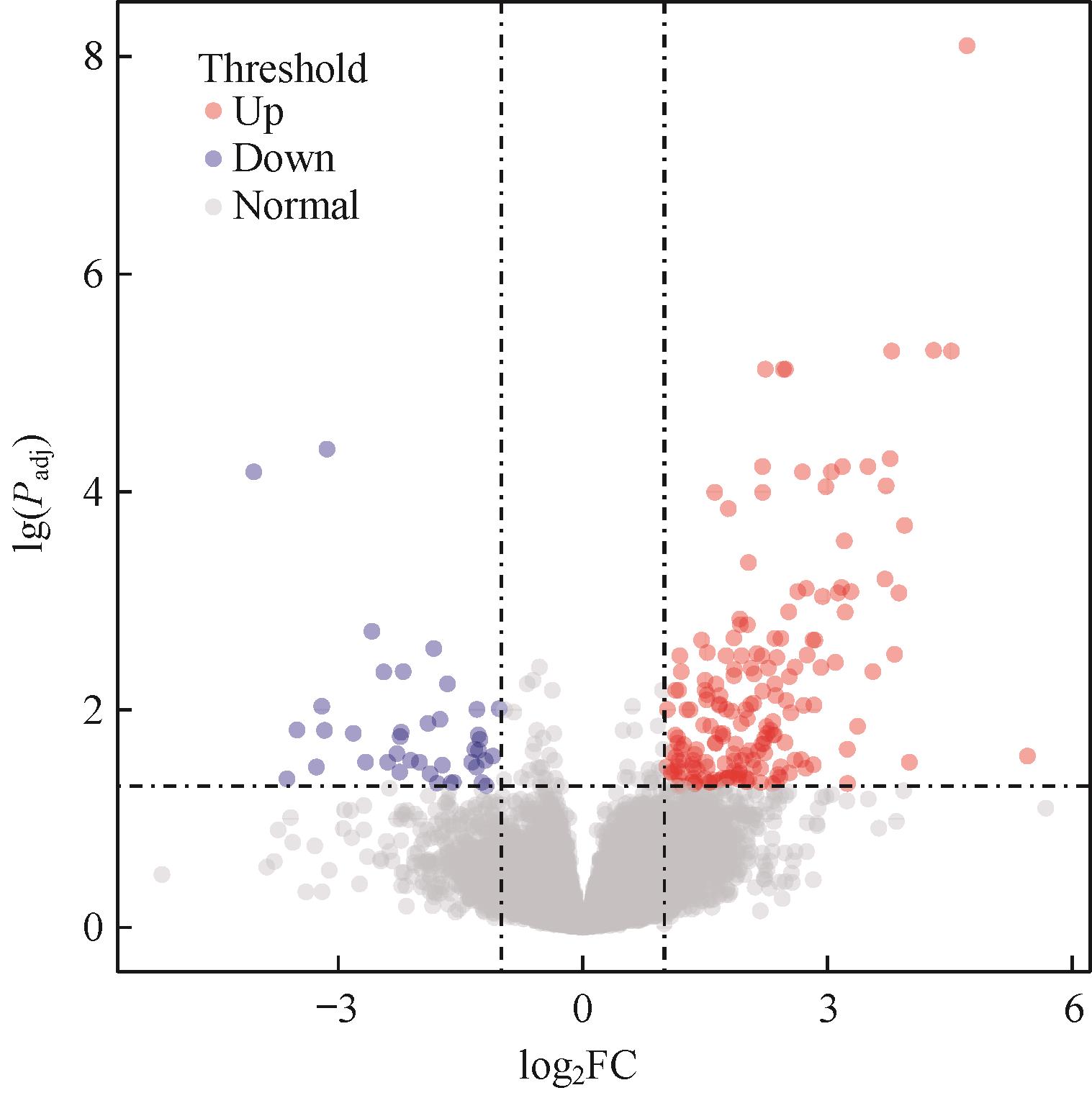
图1 MSI-H结直肠癌转移组和无转移组间DEGs火山图Note: Each dot in the graph represents a specific gene or transcript, with red dots representing significantly up-regulated genes, blue dots representing significantly down-regulated genes, and gray dots representing non-significantly different genes.
Fig 1 Volcano plot of DEGs between metastatic group and non-metastatic group of MSI-H colorectal cancer
| DEG | Full name of gene | FDR | log2 FC |
|---|---|---|---|
| Up-regulated | |||
| CA1 | Carbonic anhydrase 1 | 5.85×10-10 | 5.012 |
| IGLJ2 | Immunoglobulin lambda joining 2 | 8.21×10-6 | 4.686 |
| MS4A12 | Membrane spanning 4-domains A12 | 3.68×10-6 | 4.622 |
| SST | Somatostatin | 9.04×10-7 | 4.547 |
| GCG | Glucagon | 1.58×10-3 | 3.987 |
| SLC26A3 | Solute carrier family 26 member 3 | 2.87×10-5 | 3.960 |
| IGKV3OR2-5 | Immunoglobulin κ variable 3 or 2-5 (pseudogene) | 4.42×10-3 | 3.901 |
| OGDHL | Oxoglutarate dehydrogenase l | 1.53×10-5 | 3.878 |
| AQP8 | Aquaporin 8 | 4.66×10-4 | 3.828 |
| HTR3C | 5-hydroxytryptamine receptor 3C | 6.22×10-3 | 3.813 |
| Down-regulated | |||
| GP2 | Glycoprotein 2 | 8.44×10-5 | -4.070 |
| UICLM | Up-regulated in colorectal cancer liver metastasis | 3.59×10-2 | -3.663 |
| FGL1 | Fibrinogen like 1 | 1.08×10-2 | -3.245 |
| GPRC6A | G protein-coupled receptor class C group 6 member A | 1.91×10-2 | -3.231 |
| DEFA6 | Defensin α 6 | 3.69×10-2 | -3.231 |
| ACTL8 | Actin like 8 | 6.77×10-5 | -3.161 |
| HSD3B1 | Hydroxy-δ-5-steroid dehydrogenase, 3 β- and steroid δ-isomerase 1 | 2.12×10-2 | -2.789 |
| PADI3 | Peptidyl arginine deiminase 3 | 2.72×10-2 | -2.509 |
| MRLN | Myoregulin | 3.02×10-2 | -2.477 |
| SLC9A4 | Solute carrier family 9 member A4 | 2.03×10-2 | -2.344 |
表2 转移组和无转移组间上调及下调前10位DEGs
Tab 2 Top 10 up-regulated and down-regulated DEGs between metastasis and non-metastasis group
| DEG | Full name of gene | FDR | log2 FC |
|---|---|---|---|
| Up-regulated | |||
| CA1 | Carbonic anhydrase 1 | 5.85×10-10 | 5.012 |
| IGLJ2 | Immunoglobulin lambda joining 2 | 8.21×10-6 | 4.686 |
| MS4A12 | Membrane spanning 4-domains A12 | 3.68×10-6 | 4.622 |
| SST | Somatostatin | 9.04×10-7 | 4.547 |
| GCG | Glucagon | 1.58×10-3 | 3.987 |
| SLC26A3 | Solute carrier family 26 member 3 | 2.87×10-5 | 3.960 |
| IGKV3OR2-5 | Immunoglobulin κ variable 3 or 2-5 (pseudogene) | 4.42×10-3 | 3.901 |
| OGDHL | Oxoglutarate dehydrogenase l | 1.53×10-5 | 3.878 |
| AQP8 | Aquaporin 8 | 4.66×10-4 | 3.828 |
| HTR3C | 5-hydroxytryptamine receptor 3C | 6.22×10-3 | 3.813 |
| Down-regulated | |||
| GP2 | Glycoprotein 2 | 8.44×10-5 | -4.070 |
| UICLM | Up-regulated in colorectal cancer liver metastasis | 3.59×10-2 | -3.663 |
| FGL1 | Fibrinogen like 1 | 1.08×10-2 | -3.245 |
| GPRC6A | G protein-coupled receptor class C group 6 member A | 1.91×10-2 | -3.231 |
| DEFA6 | Defensin α 6 | 3.69×10-2 | -3.231 |
| ACTL8 | Actin like 8 | 6.77×10-5 | -3.161 |
| HSD3B1 | Hydroxy-δ-5-steroid dehydrogenase, 3 β- and steroid δ-isomerase 1 | 2.12×10-2 | -2.789 |
| PADI3 | Peptidyl arginine deiminase 3 | 2.72×10-2 | -2.509 |
| MRLN | Myoregulin | 3.02×10-2 | -2.477 |
| SLC9A4 | Solute carrier family 9 member A4 | 2.03×10-2 | -2.344 |
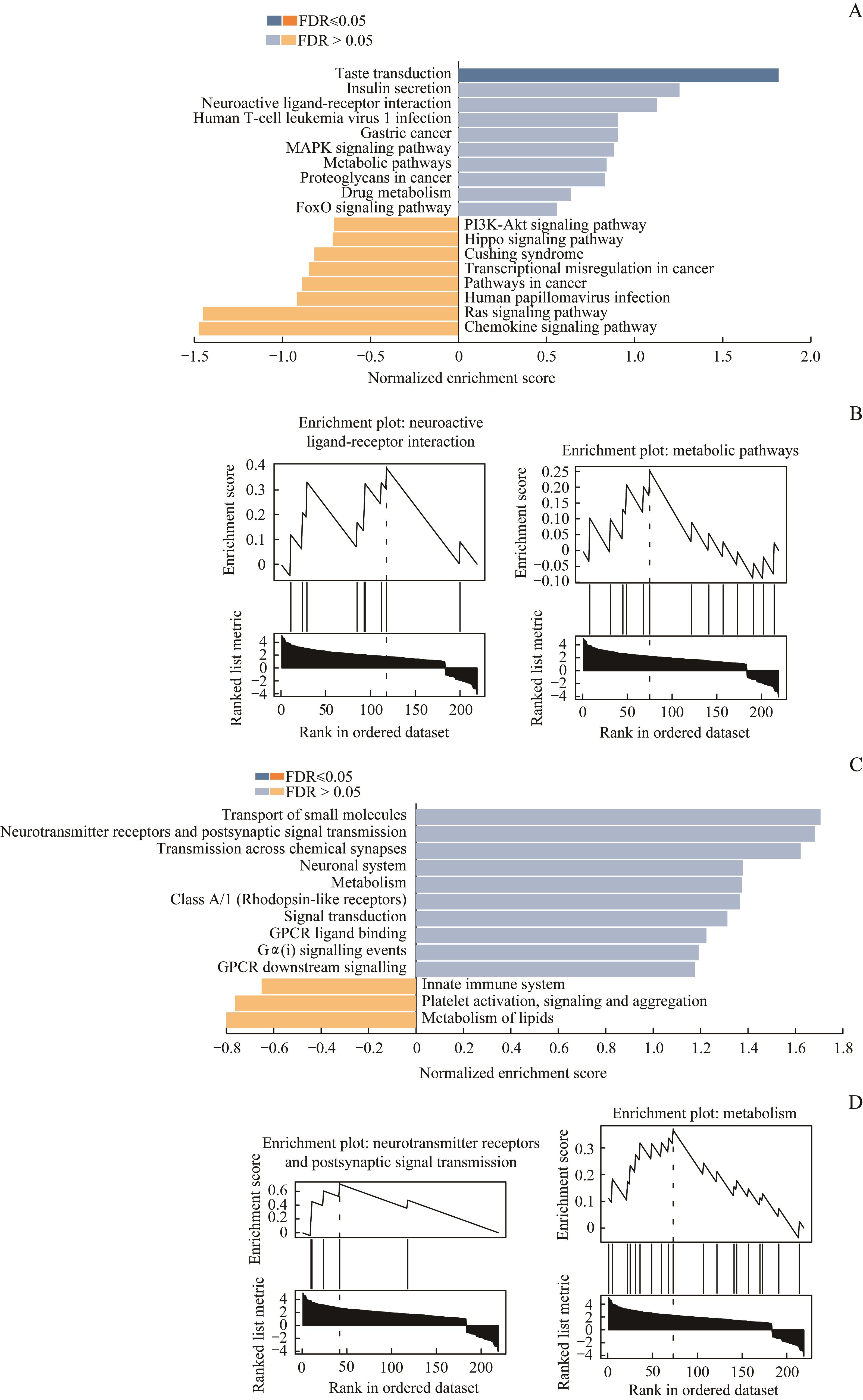
图3 DEG信号通路的GSEANote: A. The respective top 10 pathways of up-regulated genes and down-regulated genes were enriched by GSEA in KEGG database. B. Neuroactive ligand-receptor interaction and metabolic pathways were the same two pathways enriched in KEGG database and Reactome database. C. The respective top 10 pathways of up-regulated genes and down-regulated genes were enriched by GSEA in Reactome database. D. Neurotransmitter receptors and postsynaptic signal transmission and metabolism were the same two pathways enriched in Reactome database and KEGG database.
Fig 3 GSEA of DEG pathway
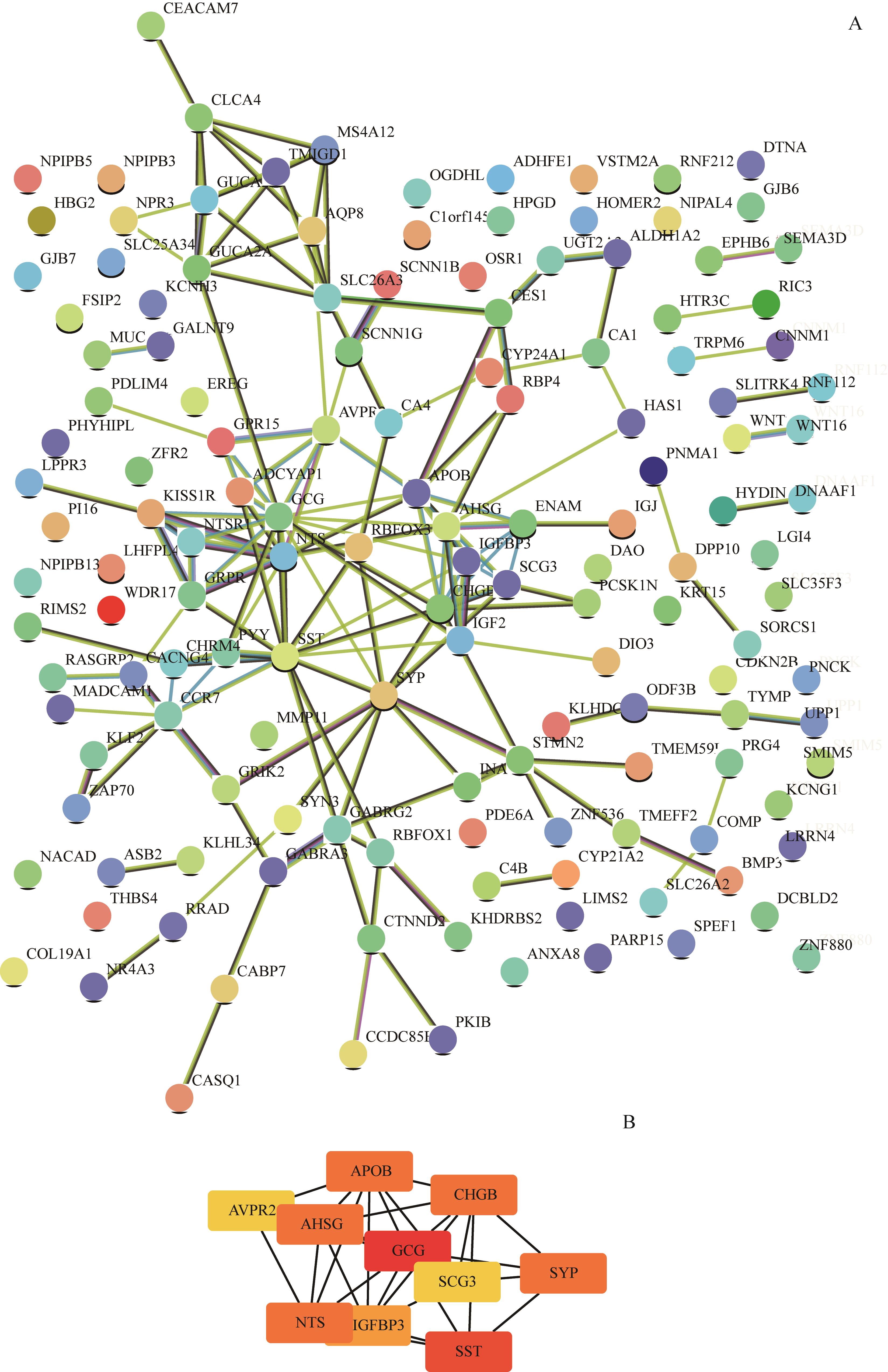
图4 上调基因PPI网络及等级前10位的hub基因Note: A. The PPI network of the up-regulated genes was constructed by STRING. B. The top 10 hub genes were analysed by Cytoscape.
Fig 4 PPI network of the up-regulated genes and the top 10 hub genes
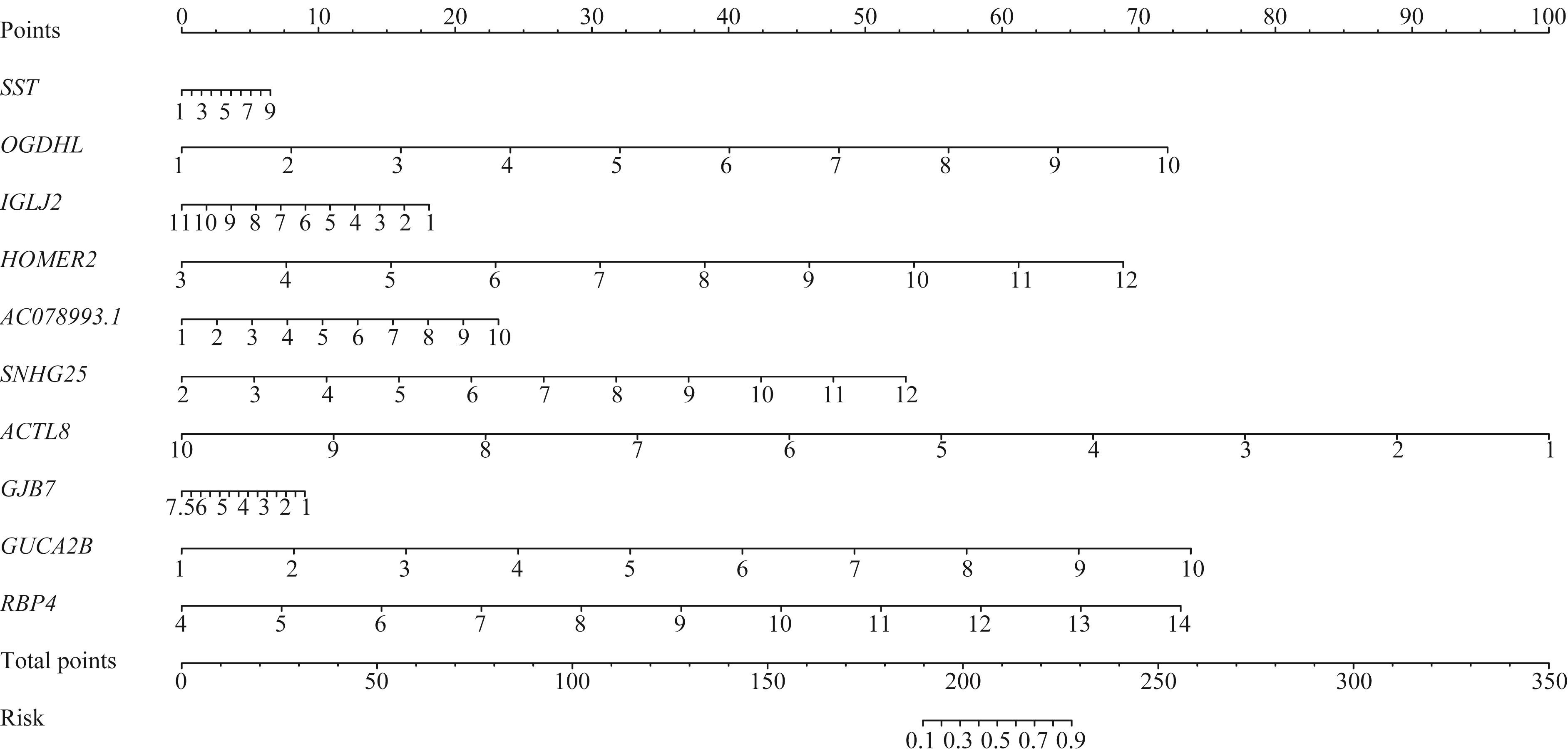
图5 MSI-H结直肠癌转移风险列线图模型Note: The value of log2∣FC∣ of the gene corresponds to the relative value on the gene scale, and then corresponds to the position on “point” scale to get the relative score. The sum of all the scores corresponds to the relative value on “total points” scale, and then corresponds to the position on “risk” scale to get the relative metastatic risk value. HOMER2—homer scaffold protein 2; SNHG25—small nucleolar RNA host gene 25; GJB7—gap junction protein beta 7.
Fig 5 Nomogram model of MSI-H colorectal cancer metastatic risk
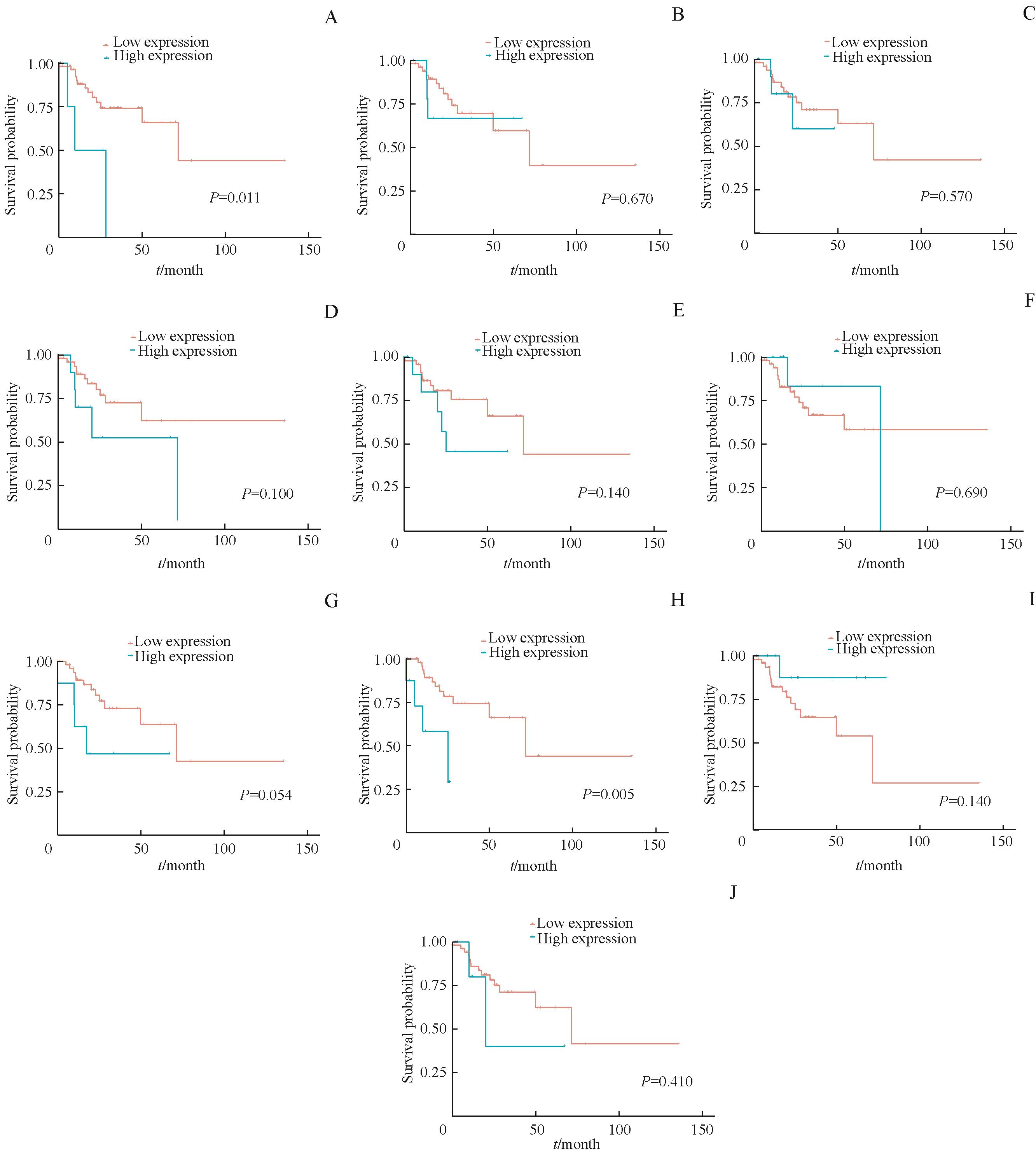
图6 生存分析列线图中10个基因对MSI-H结直肠癌PFS影响Note: The influence of the expression level of AC078993.1 (A), GJB7 (B), HOMER2 (C), OGDHL (D), SNHG25 (E), ACTL8 (F), GUCA2B (G), IGLJ2 (H), RBP4 (I) and SST (J) on PFS in MSI-H CRC.
Fig 6 Survival analysis of the 10 genes in Nomogram model on PFS of MSI-H colorectal cancer
| 1 | Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68(6): 394-424. |
| 2 | 郑荣寿, 孙可欣, 张思维, 等. 2015年中国恶性肿瘤流行情况分析[J]. 中华肿瘤杂志, 2019, 41(1): 19-28. |
| 3 | Schreuders EH, Ruco A, Rabeneck L, et al. Colorectal cancer screening: a global overview of existing programmes[J]. Gut, 2015, 64(10): 1637-1649. |
| 4 | Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017[J]. CA Cancer J Clin, 2017, 67(3): 177-193. |
| 5 | Edwards BK, Ward E, Kohler BA, et al. Annual report to the nation on the status of cancer, 1975‒2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates[J]. Cancer, 2010, 116(3): 544-573. |
| 6 | Sargent D, Sobrero A, Grothey A, et al. Evidence for cure by adjuvant therapy in colon cancer: observations based on individual patient data from 20, 898 patients on 18 randomized trials[J]. J Clin Oncol, 2009, 27(6): 872-877. |
| 7 | Copija A, Waniczek D, Witkoś A, et al. Clinical significance and prognostic relevance of microsatellite instability in sporadic colorectal cancer patients[J]. Int J Mol Sci, 2017, 18(1): E107. |
| 8 | Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence[J]. Nat Rev Clin Oncol, 2010, 7(3): 153-162. |
| 9 | Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer[J]. Cancer Res, 1998, 58(22): 5248-5257. |
| 10 | Cohen R, Svrcek M, Dreyer C, et al. New therapeutic opportunities based on DNA mismatch repair and BRAF status in metastatic colorectal cancer[J]. Curr Oncol Rep, 2016, 18(3): 18. |
| 11 | Latham A, Srinivasan P, Kemel Y, et al. Microsatellite instability is associated with the presence of lynch syndrome pan-cancer[J]. J Clin Oncol, 2019, 37(4): 286-295. |
| 12 | Søreide K, Nedrebø BS, Søreide JA, et al. Lymph node harvest in colon cancer: influence of microsatellite instability and proximal tumor location[J]. World J Surg, 2009, 33(12): 2695-2703. |
| 13 | Buckowitz A, Knaebel HP, Benner A, et al. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases[J]. Br J Cancer, 2005, 92(9): 1746-1753. |
| 14 | Malesci A, Laghi L, Bianchi P, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer[J]. Clin Cancer Res, 2007, 13(13): 3831-3839. |
| 15 | Kim CG, Ahn JB, Jung M, et al. Effects of microsatellite instability on recurrence patterns and outcomes in colorectal cancers[J]. Br J Cancer, 2016, 115(1): 25-33. |
| 16 | Venderbosch S, Nagtegaal ID, Maughan TS, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies[J]. Clin Cancer Res, 2014, 20(20): 5322-5330. |
| 17 | Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer[J]. Curr Treat Options Oncol, 2015, 16(7): 30. |
| 18 | Colle R, Cohen R, Cochereau D, et al. Immunotherapy and patients treated for cancer with microsatellite instability[J]. Bull Cancer, 2017, 104(1): 42-51. |
| 19 | Liu Y, Sethi NS, Hinoue T, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas[J]. Cancer Cell, 2018, 33(4): 721-735.e8. |
| 20 | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data[J]. Bioinformatics, 2010, 26(1): 139-140. |
| 21 | Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13(11): 2498-2504. |
| 22 | Wang SD, Yang L, Ci B, et al. Development and validation of a nomogram prognostic model for SCLC patients[J]. J Thorac Oncol, 2018, 13(9): 1338-1348. |
| 23 | Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer[J]. N Engl J Med, 2009, 361(25): 2449-2460. |
| 24 | Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis[J]. J Clin Oncol, 2005, 23(3): 609-618. |
| 25 | Koslowski M, Türeci O, Huber C, et al. Selective activation of tumor growth-promoting Ca2+channel MS4A12 in colon cancer by caudal type homeobox transcription factor CDX2[J]. Mol Cancer, 2009, 8: 77. |
| 26 | Bie FL, Wang GH, Qu X, et al. Loss of FGL1 induces epithelial‑mesenchymal transition and angiogenesis in LKB1 mutant lung adenocarcinoma[J]. Int J Oncol, 2019, 55(3): 697-707. |
| 27 | Nayeb-Hashemi H, Desai A, Demchev V, et al. Targeted disruption of fibrinogen like protein-1 accelerates hepatocellular carcinoma development[J]. Biochem Biophys Res Commun, 2015, 465(2): 167-173. |
| 28 | Chai ZB, Wang L, Zheng YB, et al. PADI3 plays an antitumor role via the Hsp90/CKS1 pathway in colon cancer[J]. Cancer Cell Int, 2019, 19: 277. |
| 29 | Chang XT, Chai ZB, Zou JR, et al. PADI3 induces cell cycle arrest via the Sirt2/AKT/p21 pathway and acts as a tumor suppressor gene in colon cancer[J]. Cancer Biol Med, 2019, 16(4): 729-742. |
| 30 | Prevarskaya N, Skryma R, Shuba Y. Ion channels in cancer: are cancer hallmarks oncochannelopathies?[J]. Physiol Rev, 2018, 98(2): 559-621. |
| 31 | Lui VC, Lung SS, Pu JK, et al. Invasion of human glioma cells is regulated by multiple chloride channels including ClC-3[J]. Anticancer Res, 2010, 30(11): 4515-4524. |
| 32 | Siveen KS, Raza A, Ahmed EI, et al. The role of extracellular vesicles as modulators of the tumor microenvironment, metastasis and drug resistance in colorectal cancer[J]. Cancers (Basel), 2019, 11(6): E746. |
| 33 | la Vecchia S, Sebastián C. Metabolic pathways regulating colorectal cancer initiation and progression[J]. Semin Cell Dev Biol, 2020, 98: 63-70. |
| 34 | Kasprzak A, Adamek A. The neuropeptide system and colorectal cancer liver metastases: mechanisms and management[J]. Int J Mol Sci, 2020, 21(10): 3494. |
| 35 | Qiu SY, Nikolaou S, Zhu J, et al. Characterisation of the expression of neurotensin and its receptors in human colorectal cancer and its clinical implications[J]. Biomolecules, 2020, 10(8): 1145. |
| 36 | Liu Y, He JJ, Xu JH, et al. Neuroendocrine differentiation is predictive of poor survival in patients with stage Ⅱ colorectal cancer[J]. Oncol Lett, 2017, 13(4): 2230-2236. |
| 37 | Yamamoto N, Oshima T, Yoshihara K, et al. Clinicopathological significance and impact on outcomes of the gene expression levels of IGF-1, IGF-2 and IGF-1R, IGFBP-3 in patients with colorectal cancer: overexpression of the IGFBP-3 gene is an effective predictor of outcomes in patients with colorectal cancer[J]. Oncol Lett, 2017, 13(5): 3958-3966. |
| 38 | Yang LS, Li JY, Fu SZ, et al. Up-regulation of insulin-like growth factor binding protein-3 is associated with brain metastasis in lung adenocarcinoma[J]. Mol Cells, 2019, 42(4): 321-332. |
| 39 | Loboda A, Nebozhyn MV, Watters JW, et al. EMT is the dominant program in human colon cancer[J]. BMC Med Genomics, 2011, 4: 9. |
| 40 | Schell MJ, Yang ML, Missiaglia E, et al. A composite gene expression signature optimizes prediction of colorectal cancer metastasis and outcome[J]. Clin Cancer Res, 2016, 22(3): 734-745. |
| [1] | 李偲羽, 陈娅, 胡文韬, 戴勇鸣, 吴颖为. 磁共振耦合谱成像识别头颈部肿瘤异质性及隐匿性淋巴结转移[J]. 上海交通大学学报(医学版), 2025, 45(9): 1202-1213. |
| [2] | 江怡, 黄晨浩, 李祉良, 吴珺玮, 赵任, 张弢. 1例KRAS突变的结直肠癌患者术前接受化疗联合免疫治疗的效果报道[J]. 上海交通大学学报(医学版), 2025, 45(9): 1256-1260. |
| [3] | 吴雷, 杜凤麟, 赵明娜, 任逸喆, 张先洲, 娄加陶. N型蛋白酪氨酸磷酸酶受体在肺腺癌中的表达及其促进肿瘤转移的机制[J]. 上海交通大学学报(医学版), 2025, 45(7): 846-857. |
| [4] | 王蕊, 袁瑛, 陶晓峰. 合成磁共振成像在口腔癌颈部淋巴结转移诊断中的应用价值[J]. 上海交通大学学报(医学版), 2025, 45(7): 900-909. |
| [5] | 杨娜, 刘俊丽, 白静, 杨思怡, 韩继明, 张华华. HENMT1通过激活PI3K-AKT-mTOR信号通路促进胃癌的增殖与迁移[J]. 上海交通大学学报(医学版), 2025, 45(6): 717-726. |
| [6] | 张钲佳, 李小敏, 周鑫, 马海荣, 艾松涛. 高阶磁共振功能成像评估骨与软组织肿瘤价值初探[J]. 上海交通大学学报(医学版), 2025, 45(5): 585-596. |
| [7] | 陈蓉, 张锰, 朱荻绮, 郭颖, 沈捷. 基于抗中性粒细胞胞质抗体的列线图模型对川崎病患儿并发冠状动脉病变风险的预测作用[J]. 上海交通大学学报(医学版), 2025, 45(4): 459-467. |
| [8] | 邓佳丽, 郭嘉婧, 王静怡, 丁心怡, 朱仪, 王中领. 自组装载药纳米探针用于乳腺癌焦亡增敏及化学交换饱和转移成像研究[J]. 上海交通大学学报(医学版), 2025, 45(3): 271-281. |
| [9] | 陈佳莹, 褚以忞, 彭海霞. 结直肠癌无进展生存时间预测模型及影响因素研究[J]. 上海交通大学学报(医学版), 2025, 45(3): 324-334. |
| [10] | 刘楚萱, 左佳鑫, 熊屏. 基于超声评分参数及临床指标的列线图鉴别原发性干燥综合征与IgG4相关唾液腺炎[J]. 上海交通大学学报(医学版), 2025, 45(3): 373-380. |
| [11] | 梁乐斌, 陈慧芳, 赖淑静, 顾靓, 苏冰. 基于空间ATAC-seq技术的Apcmin/+小鼠结肠肿瘤表观特征分析[J]. 上海交通大学学报(医学版), 2025, 45(10): 1261-1270. |
| [12] | 张先洲, 杜凤麟, 吴雷, 任逸喆, 赵明娜, 娄加陶. OGT通过ERK信号通路促进非小细胞肺癌增殖的机制研究[J]. 上海交通大学学报(医学版), 2025, 45(10): 1288-1297. |
| [13] | 禹志远, 董海平, 高楠, 马柯. 背根神经节吗啡耐受核心基因筛选与机制研究:加权基因共表达网络分析和机器学习的转录组学整合策略[J]. 上海交通大学学报(医学版), 2025, 45(10): 1308-1319. |
| [14] | 敦译霆, 赵婧, 冯成领, 李行健, 崔迪, 韩邦旻. 机器人辅助腹腔镜根治性前列腺切除术后患者尿失禁的在线风险计算器和列线图预测模型[J]. 上海交通大学学报(医学版), 2025, 45(10): 1361-1371. |
| [15] | 陆佳萍, 刘醒, 张林杉, 赵琳, 张敏, 李小英, 刘玥隽. 腹部脂肪面积与2型糖尿病患者胰岛β细胞第一时相分泌功能的关系[J]. 上海交通大学学报(医学版), 2025, 45(1): 42-50. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||