
上海交通大学学报(医学版) ›› 2022, Vol. 42 ›› Issue (11): 1589-1597.doi: 10.3969/j.issn.1674-8115.2022.11.011
收稿日期:2022-05-07
接受日期:2022-10-18
出版日期:2022-11-28
发布日期:2023-01-04
通讯作者:
郭润生,电子信箱: grs982600@163.com。*为共同通信作者。作者简介:孙亚蒙(1986—),男,博士,电子信箱: symbuster_1986@163.com。第一联系人:(马晔、郭润生并列第一作者)
基金资助:
SUN Yameng1( ), MA Ye2(
), MA Ye2( ), GUO Runsheng2(
), GUO Runsheng2( )
)
Received:2022-05-07
Accepted:2022-10-18
Online:2022-11-28
Published:2023-01-04
Contact:
GUO Runsheng, E-mail: grs982600@163.com. *Co-corresponding authors.Supported by:摘要:
目的·利用微滴式数字PCR系统(droplet digital PCR system,ddPCR)建立乳腺癌组织福尔马林固定石蜡包埋(formalin-fixed paraffin embedded,FFPE)样本和血浆样本中 HER2基因拷贝数检测体系,并对检测体系进行性能评估,为临床辅助诊疗提供科学依据。方法·收集2020年1月—2021年6月就诊于上海健康医学院附属嘉定区中心医院普外科乳腺癌患者组织样本12例及其中4例配对血浆样本用于检测体系评价,同期收集健康志愿者血浆样本24例及就诊的乳腺癌组织切片样本77例,提取核酸,用于建立检测体系空白检测限及性能评价。设计及筛选检测体系,使用ddPCR检测体系的空白检测限、精密度、灵敏度及线性范围,使用高通量测序(next generation sequencing,NGS)验证ddPCR检测一致性,并与荧光原位杂交(fluorescence in situ hybridization,IHC)/免疫组化(immunohistochemistry,FISH)金标准方法比对检测体系准确性。定量资料比较采用 χ 2 检验和配对样本 t检验进行分析,使用Poisson概率函数分析体系空白检测限,变异系数(coefficient of variation,CV)评价灵敏度及精密度,线性回归相关系数( R 2 )评价线性范围。结果·使用ddPCR平台建立了乳腺癌患者组织切片DNA中 HER2基因拷贝数定量和扩增检测体系,配对样本 t检验评价:与NGS方法检测组织核酸和血浆游离核酸样本结果相比,检测拷贝数结果相关系数0.987,扩增判读一致性100%。评估检测体系的批内精密度为6.8%、批间精密度为9.4%,灵敏度为1 ng,线性良好( R 2 >0.98)。与传统的IHC/FISH方法相比,检测体系的灵敏度和特异度分别为84.6%(95% CI 64.3%~95.0%)和76.5%(95% CI 62.2%~86.8%), Kappa为0.57,一致率为79.2%。结论·使用ddPCR建立乳腺癌患者 HER2拷贝数检测体系,临床检测一致性良好,可能为乳腺癌患者的诊疗和预后判断的研究提供辅助手段。
中图分类号:
孙亚蒙, 马晔, 郭润生. 乳腺癌组织样本中 HER2基因拷贝数的检测分析[J]. 上海交通大学学报(医学版), 2022, 42(11): 1589-1597.
SUN Yameng, MA Ye, GUO Runsheng. Detection and analysis of copy number of HER2 gene in breast cancer tissue samples[J]. Journal of Shanghai Jiao Tong University (Medical Science), 2022, 42(11): 1589-1597.
| Gene | Forword primer (5′→3′) | Reverse primer (5′→3′) | Probe sequence (5′→3′) |
|---|---|---|---|
| HER2 | TCACTCATATCCTCCTCTTTCTGC | AATTTTCACATTCTCCCCATCAG | FAM-CAGGGCATCTGGATC-MGB |
| EIF2C1 | GCTGCTAGGCTTTCCTGTTC | GCCTATTTTCCTGCATCTTCT | HEX-AGGCCCCAAAACCCTAAAC-MGB |
表1 引物和探针序列信息
Tab 1 List of primers and probe sequences
| Gene | Forword primer (5′→3′) | Reverse primer (5′→3′) | Probe sequence (5′→3′) |
|---|---|---|---|
| HER2 | TCACTCATATCCTCCTCTTTCTGC | AATTTTCACATTCTCCCCATCAG | FAM-CAGGGCATCTGGATC-MGB |
| EIF2C1 | GCTGCTAGGCTTTCCTGTTC | GCCTATTTTCCTGCATCTTCT | HEX-AGGCCCCAAAACCCTAAAC-MGB |
| Sample | Detection System 1 | BC-HER2-2019 | ||||
|---|---|---|---|---|---|---|
| HER2 gene copy number (copies) | Reference gene copy number (copies) | Copy amplification [( HER2/Reference)]×2 | HER2 gene copy number (copies) | Reference gene copy number (copies) | Copy amplification [( HER2/Reference)]×2 | |
| HV-1 | 2 400 | 1.33 | 3 180 | 1.99 | ||
| HV-2 | 2 150 | 1.25 | 3 050 | 1.97 | ||
| HV-3 | 2 600 | 1.41 | 3 200 | 2.05 | ||
表2 健康人 cfDNA中 HER2 基因和内参基因拷贝数及拷贝数比例
Tab 2 Copy number and copy number ratio of HER2 and reference gene in healthy human cfDNA
| Sample | Detection System 1 | BC-HER2-2019 | ||||
|---|---|---|---|---|---|---|
| HER2 gene copy number (copies) | Reference gene copy number (copies) | Copy amplification [( HER2/Reference)]×2 | HER2 gene copy number (copies) | Reference gene copy number (copies) | Copy amplification [( HER2/Reference)]×2 | |
| HV-1 | 2 400 | 1.33 | 3 180 | 1.99 | ||
| HV-2 | 2 150 | 1.25 | 3 050 | 1.97 | ||
| HV-3 | 2 600 | 1.41 | 3 200 | 2.05 | ||
| Sample No. | Concentration/(ng·μL -1) | HER2 Gene copy | Sample No. | Concentration/(ng·μL -1) | HER2 Gene copy |
|---|---|---|---|---|---|
| 1 | 0.44 | 1.80 | 13 | 0.21 | 2.24 |
| 2 | 0.19 | 2.20 | 14 | 0.10 | 2.20 |
| 3 | 0.54 | 2.08 | 15 | 0.16 | 2.22 |
| 4 | 0.26 | 2.01 | 16 | 0.10 | 1.20 |
| 5 | 0.95 | 2.58 | 17 | 0.38 | 2.37 |
| 6 | 0.62 | 2.00 | 18 | 0.13 | 1.87 |
| 7 | 0.23 | 2.90 | 19 | 0.49 | 1.99 |
| 8 | 0.29 | 2.27 | 20 | 0.32 | 1.62 |
| 9 | 0.32 | 1.99 | 21 | 0.10 | 2.40 |
| 10 | 0.21 | 2.28 | 22 | 0.24 | 1.75 |
| 11 | 0.23 | 2.08 | 23 | 0.19 | 2.19 |
| 12 | 0.38 | 2.17 | 24 | 0.74 | 1.69 |
表3 24例健康人 cfDNA样本 HER2拷贝数比例
Tab 3 HER2 copy number ratio in 24 healthy human cfDNA samples
| Sample No. | Concentration/(ng·μL -1) | HER2 Gene copy | Sample No. | Concentration/(ng·μL -1) | HER2 Gene copy |
|---|---|---|---|---|---|
| 1 | 0.44 | 1.80 | 13 | 0.21 | 2.24 |
| 2 | 0.19 | 2.20 | 14 | 0.10 | 2.20 |
| 3 | 0.54 | 2.08 | 15 | 0.16 | 2.22 |
| 4 | 0.26 | 2.01 | 16 | 0.10 | 1.20 |
| 5 | 0.95 | 2.58 | 17 | 0.38 | 2.37 |
| 6 | 0.62 | 2.00 | 18 | 0.13 | 1.87 |
| 7 | 0.23 | 2.90 | 19 | 0.49 | 1.99 |
| 8 | 0.29 | 2.27 | 20 | 0.32 | 1.62 |
| 9 | 0.32 | 1.99 | 21 | 0.10 | 2.40 |
| 10 | 0.21 | 2.28 | 22 | 0.24 | 1.75 |
| 11 | 0.23 | 2.08 | 23 | 0.19 | 2.19 |
| 12 | 0.38 | 2.17 | 24 | 0.74 | 1.69 |
| Sample No. | Test date | Copy amplification [( HER2/Reference)]×2 | |||||
|---|---|---|---|---|---|---|---|
| Batch 1 | Batch 2 | Batch 3 | |||||
| Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | ||
| 1 | Day 1 | 1.74 | 2.14 | 2.16 | 2.14 | 1.82 | 1.94 |
| 2 | Day 2 | 1.84 | 1.88 | 2.36 | 2.18 | 1.92 | 1.88 |
| 3 | Day 3 | 2.08 | 1.98 | 2.38 | 2.50 | 2.20 | 1.82 |
| 4 | Day 4 | 2.12 | 2.26 | 1.83 | 2.14 | 1.80 | 1.90 |
| 5 | Day 5 | 2.10 | 2.10 | 2.24 | 2.04 | 2.18 | 1.92 |
| Xmean | 2.05 | ||||||
| Intra batch standard deviation | 0.14 | ||||||
| inter batch standard deviation | 0.19 | ||||||
| Intra batch precision/% | 6.80 | ||||||
| inter batch precision/% | 9.40 | ||||||
表4 检测体系的精密度评价
Tab 4 Precision evaluation of the detection system
| Sample No. | Test date | Copy amplification [( HER2/Reference)]×2 | |||||
|---|---|---|---|---|---|---|---|
| Batch 1 | Batch 2 | Batch 3 | |||||
| Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | Repeat 1 | Repeat 2 | ||
| 1 | Day 1 | 1.74 | 2.14 | 2.16 | 2.14 | 1.82 | 1.94 |
| 2 | Day 2 | 1.84 | 1.88 | 2.36 | 2.18 | 1.92 | 1.88 |
| 3 | Day 3 | 2.08 | 1.98 | 2.38 | 2.50 | 2.20 | 1.82 |
| 4 | Day 4 | 2.12 | 2.26 | 1.83 | 2.14 | 1.80 | 1.90 |
| 5 | Day 5 | 2.10 | 2.10 | 2.24 | 2.04 | 2.18 | 1.92 |
| Xmean | 2.05 | ||||||
| Intra batch standard deviation | 0.14 | ||||||
| inter batch standard deviation | 0.19 | ||||||
| Intra batch precision/% | 6.80 | ||||||
| inter batch precision/% | 9.40 | ||||||
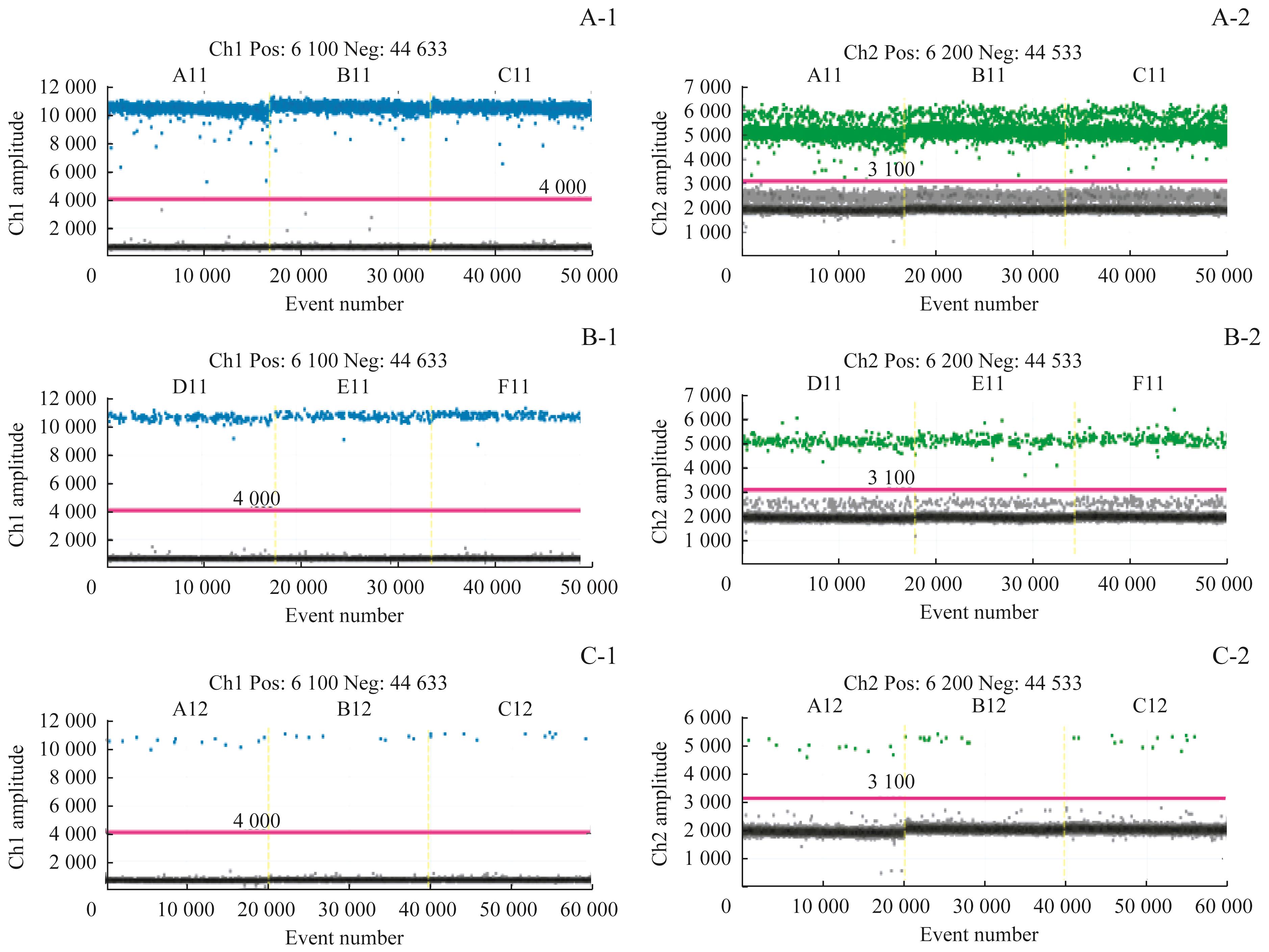
图2 灵敏度检测结果
Fig 2 Results of sensitivity detectionNote: A-1/A-2. Results of three duplication holes of HER2 and reference gene by using 10ng. B-1/B-2. Results of three duplication holes of HER2 and reference gene by using 1ng. C-1/C-2. Results of three duplication holes of HER2 and reference gene by using 0.1ng.
| Loading quantity | Repeat | HER2 Gene copy number (copies/20 μL) | Reference Gene copy number (copies/20 μL) | Copy amplification [( HER2/Reference)]×2 | CV/% |
|---|---|---|---|---|---|
| 10 ng | 1 | 2 780 | 1.89 | 3.30 | |
| 2 | 3 100 | 1.99 | |||
| 3 | 3 160 | 2.01 | |||
| 1 ng | 1 | 226 | 274 | 1.64 | 12.50 |
| 2 | 234 | 244 | 1.91 | ||
| 3 | 276 | 260 | 2.11 | ||
| 0.1 ng | 1 | 15.20 | 14.00 | 2.20 | 26.00 |
| 2 | 9.60 | 14.40 | 1.30 | ||
| 3 | 13.00 | 15.40 | 1.70 |
表5 灵敏度检测拷贝数统计及变异系数
Tab 5 Copy number statistics and CV of sensitivity detection
| Loading quantity | Repeat | HER2 Gene copy number (copies/20 μL) | Reference Gene copy number (copies/20 μL) | Copy amplification [( HER2/Reference)]×2 | CV/% |
|---|---|---|---|---|---|
| 10 ng | 1 | 2 780 | 1.89 | 3.30 | |
| 2 | 3 100 | 1.99 | |||
| 3 | 3 160 | 2.01 | |||
| 1 ng | 1 | 226 | 274 | 1.64 | 12.50 |
| 2 | 234 | 244 | 1.91 | ||
| 3 | 276 | 260 | 2.11 | ||
| 0.1 ng | 1 | 15.20 | 14.00 | 2.20 | 26.00 |
| 2 | 9.60 | 14.40 | 1.30 | ||
| 3 | 13.00 | 15.40 | 1.70 |
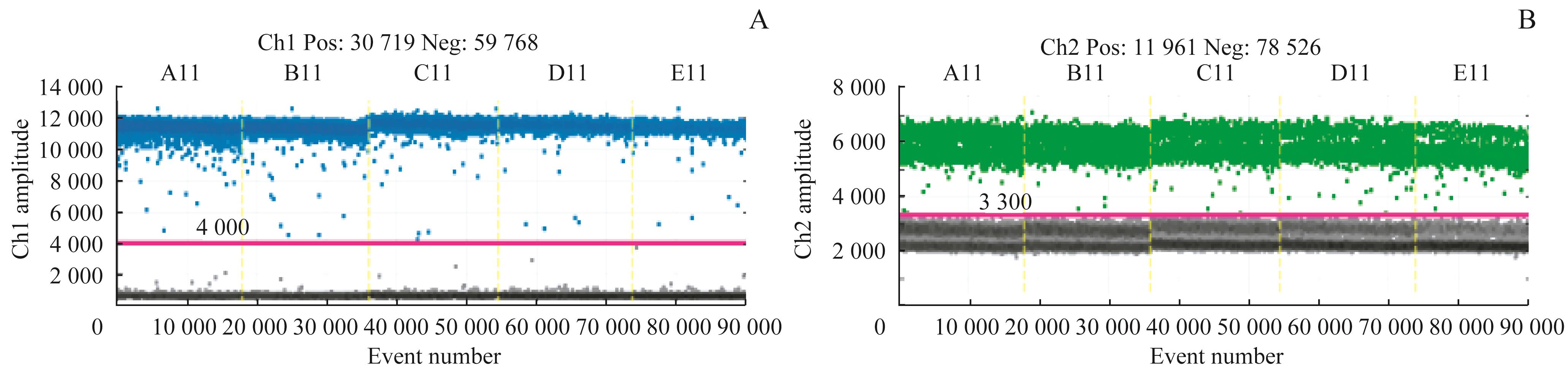
图3 Her2 基因和内参基因的 ddPCR线性检测扩增图谱Note: A. The result of the HER2 copies. B. The result of the reference gene copies.
Fig 3 Linear detection amplification plots of the gene HER2 and reference by ddPCR
HER2∶ reference | Theoretical value | Actual value | |||||
|---|---|---|---|---|---|---|---|
HER2 gene copy number (copies/20 μL) | Reference gene copy number (copies/20 μL) | Copy amplification [( HER2/ reference)]×2 | HER2 gene copy number (copies/20 μL) | Reference gene copy number (copies/20 μL) | Copy amplification [( HER2/ reference)]×2 | ||
| 1∶1 | 3 260 | 2.04 | 3 160 | 1.89 | |||
| 2∶1 | 5 984 | 3.93 | 6 460 | 2.99 | |||
| 3∶1 | 8 708 | 6.01 | 8 900 | 3 000 | 5.94 | ||
| 4∶1 | 13 914 | 7.96 | 16 140 | 8.62 | |||
| 5∶1 | 14 482 | 2927 | 9.90 | 16 760 | 10.00 | ||
表6 线性检测拷贝数统计
Tab 6 Copy number statistics of linear detection
HER2∶ reference | Theoretical value | Actual value | |||||
|---|---|---|---|---|---|---|---|
HER2 gene copy number (copies/20 μL) | Reference gene copy number (copies/20 μL) | Copy amplification [( HER2/ reference)]×2 | HER2 gene copy number (copies/20 μL) | Reference gene copy number (copies/20 μL) | Copy amplification [( HER2/ reference)]×2 | ||
| 1∶1 | 3 260 | 2.04 | 3 160 | 1.89 | |||
| 2∶1 | 5 984 | 3.93 | 6 460 | 2.99 | |||
| 3∶1 | 8 708 | 6.01 | 8 900 | 3 000 | 5.94 | ||
| 4∶1 | 13 914 | 7.96 | 16 140 | 8.62 | |||
| 5∶1 | 14 482 | 2927 | 9.90 | 16 760 | 10.00 | ||
| No. | NGS copy amplification | ddPCR copy amplification |
|---|---|---|
| 1 a | 1.90 | 1.62 |
| 2 a | 1.90 | 1.86 |
| 3 a | 1.90 | 1.62 |
| 4 a | 5.00 | 4.27 |
| 5 a | 6.00 | 4.21 |
| 6 a | 13.10 | 14.10 |
| 7 a | 8.00 | 6.64 |
| 8 a | 18.00 | 16.00 |
| 9 a | 2.40 | 1.73 |
| 9 b | 2.10 | 1.84 |
| 10 a | 2.20 | 1.77 |
| 10 b | 1.80 | 1.41 |
| 11 a | 5.00 | 3.50 |
| 11 b | 1.80 | 1.49 |
| 12 a | 5.00 | 3.85 |
| 12 b | 1.70 | 1.20 |
表7 NGS检测拷贝数与 ddPCR检测对比
Tab 7 Copy number results by NGS comparison of ddPCR
| No. | NGS copy amplification | ddPCR copy amplification |
|---|---|---|
| 1 a | 1.90 | 1.62 |
| 2 a | 1.90 | 1.86 |
| 3 a | 1.90 | 1.62 |
| 4 a | 5.00 | 4.27 |
| 5 a | 6.00 | 4.21 |
| 6 a | 13.10 | 14.10 |
| 7 a | 8.00 | 6.64 |
| 8 a | 18.00 | 16.00 |
| 9 a | 2.40 | 1.73 |
| 9 b | 2.10 | 1.84 |
| 10 a | 2.20 | 1.77 |
| 10 b | 1.80 | 1.41 |
| 11 a | 5.00 | 3.50 |
| 11 b | 1.80 | 1.49 |
| 12 a | 5.00 | 3.85 |
| 12 b | 1.70 | 1.20 |
| ddPCR | FISH | Specificity [% (95% CI)] | Sensitivity [% (95% CI)] | Kappa value | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Positive | 22 | 12 | 76.5 (62.2—86.8) | 84.6 (64.3—95.0) | 0.57 |
| Negative | 4 | 39 | |||
表8 以 FISH为金标准评价 ddPCR方法检测 HER2 拷贝数
Tab 8 Gold standard FISH to evaluate the HER2 copy number detected by ddPCR
| ddPCR | FISH | Specificity [% (95% CI)] | Sensitivity [% (95% CI)] | Kappa value | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Positive | 22 | 12 | 76.5 (62.2—86.8) | 84.6 (64.3—95.0) | 0.57 |
| Negative | 4 | 39 | |||
| 1 | PONDÉ N, AFTIMOS P, PICCART M. Antibody-drug conjugates in breast cancer: a comprehensive review[J]. Curr Treat Options Oncol, 2019, 20(5): 37. |
| 2 | GRIGUOLO G, PASCUAL T, DIECI M V, et al. Interaction of host immunity with HER2-targeted treatment and tumor heterogeneity in HER2-positive breast cancer[J]. J Immunother Cancer, 2019, 7(1): 90. |
| 3 | MAHTANI R, HOLMES F A, BADVE S, et al. Breast Cancer Therapy Expert Group (BCTEG). A roundtable discussion of the Breast Cancer Therapy Expert Group (BCTEG): clinical developments and practice guidance on human epidermal growth factor receptor 2 ( HER2)-positive breast Cancer[J]. Clin Breast Cancer, 2020, 20(3): e251-e260. |
| 4 | WOLFF A C, HAMMOND M E H, ALLISON K H, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update[J]. J Clin Oncol, 2018, 36(20): 2105- 2122. |
| 5 | NIU D F, LI L, YU Y, et al. Evaluation of next generation sequencing for detecting HER2 copy number in breast and gastric cancers[J]. Pathol Oncol Res, 2020, 6(4): 2577-2585. |
| 6 | AHN S, WOO J W, LEE K, et al. HER2 status in breast cancer: changes in guidelines and complicating factors for interpretation[J]. J Pathol Transl Med, 2020, 54(1): 34-44. |
| 7 | 相学平. CEP17异常对评判乳腺癌 HER2基因扩增状态的影响[J]. 临床与实验病理学杂志, 2014, 30(7): 728-731. |
| XIANG X P. Effects of CEP17 abnormal on HER2 gene status evaluation in breast carcinoma[J]. Chin J Clin Exp, 2014, 30(7): 728-731. | |
| 8 | 卢仁泉, 柳光宇, 杨文涛, 等. 外周血 HER2基因扩增检测(数字PCR法)在抗 HER2治疗中的应用共识[J]. 中国癌症杂志, 2022, 32(1): 90-96. |
| LU R Q, LIU G Y, YANG W T, et al. Application consensus of peripheral blood HER2 gene amplification detection (digital PCR) in anti- HER2 therapy[J]. China Oncol, 2022, 32(1): 90-96. | |
| 9 | SHODA K, ICHIKAWA D, FUJITA Y, et al. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer[J]. Gastric Cancer, 2017, 20(1): 126-135. |
| 10 | SHODA K, MASUDA K, ICHIKAWA D, et al. HER2 amplification detected in the circulating DNA of patients with gastric cancer: a retrospective pilot study[J]. Gastric Cancer, 2015, 18(4):698-710. |
| 11 | ZHU Y Z, LU D, LIRA M E, et al. Droplet digital polymerase chain reaction detection of HER2 amplification in formalin fixed paraffin embedded breast and gastric carcinoma samples[J]. Exp Mol Pathol, 2016, 100(2): 287-293. |
| 12 | 杨文涛,步宏.乳腺癌 HER2检测指南(2019版)[J].中华病理学杂志, 2019, 48(3): 169-175. |
| YANG W T, BU H. Guidelines for detection of HER2 in breast cancer (2019 edition)[J]. Chin J Pathol, 2019, 48(3): 169-175. | |
| 13 | 陈静瑶, 周杰, 李飞, 等. 飞燕草素通过AKT/mTOR通路诱导HER-2+乳腺癌细胞自噬[J]. 中南大学学报(医学版), 2017, 42(3): 264-270. |
| CHEN S Y, ZHOU J, LI F, et al. Delphinidin induces autophagy in HER-2+ breast cancer cells via inhibition of AKT/mTOR pathway[J]. J Cent South Univ (Med Sci), 2017, 42(3): 264-270. | |
| 14 | 董周寰, 张晶, 王哲,等. 运用数字PCR检测乳腺癌组织FFPE样品中人表皮生长因子受体2拷贝数的变化[J]. 生物技术通讯, 2018, 29(3): 6. |
| DONG Z H, ZHANG J, WANG Z, et al. HER2 Copy Number Alteration in FFPE Samples Using Droplet Digital PCR[J]. Lett Biotechnol, 2018, 29(3): 6. | |
| 15 | BARTLEY A N, WASHINGTON M K, COLASACCO C, et al. HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: guideline from the College of American Pathologists, American Society for Clinical Pathology, and the American Society of Clinical Oncology [J]. J Clin Oncol, 2017, 35(4): 446-464. |
| 16 | JANJIGIAN Y Y, KAWAZOE A, YAÑEZ P, et al. The KEYNOTE-811 trial of dual PD-1 and HER2 blockade in HER2-positive gastric cancer[J]. Nature,2021, 600(7890): 727-730. |
| 17 | 曹明丽. 基于分子分型的乳腺癌流行病学新认识[J]. 中国肿瘤临床, 2017, 44(9): 449-451. |
| CAO M L. Advances in breast cancer epidemiology based on molecular subtyping[J]. Chin J Clin Oncol, 2017, 44(9): 449-451. | |
| 18 | 张艳秋, 王昳凡, 王简. HER-2阳性乳腺癌的新辅助治疗现状及展望[J]. 临床肿瘤学杂志, 2017, 22(3): 264-271. |
| ZHANG Y Q, WANG Y F, WANG J. The current and progress of neoadjuvant treatment of HER-2 positive breast cancer[J]. Chin Clin Oncol, 2017, 22(3): 264-271. | |
| 19 | VESCI L, CAROLLO V, ROSCILLI G, et al. Trastuzumab and docetaxel in a preclinical organotypic breast cancer model using tissue slices from mammary fat pad: translational relevance[J]. Oncol Rep. 2015, 34(3): 1146-1152. |
| 20 | 李明, 王涛, 杨周. 靶向药物在HER2阳性乳腺癌患者化疗中的临床应用价值研究[J]. 中国医学创新, 2022, 19(8): 51-54. |
| LI M, WANG T, YANG Z. The clinical application research of targeted drugs in chemotherapy of HER2 positive breast cancer patients[J]. Chinese Med Innovations, 2022, 19(8): 51-54. | |
| 21 | 王利锋, 袁芳, 陈锐, 等. 微滴数字PCR在检测乳腺癌 HER2基因扩增中的应用[J]. 中华病理学杂志, 2018, 47(10): 790-792. |
| WANG L F, YUAN F, CHEN R, et al. Application of droplet digital PCR technology for HER2 gene amplification of breast cancer[J]. Chin J Pathol, 2018, 47(10): 790-792. | |
| 22 | KIM B, NAM S K, SEO S H, et al. Comparative analysis of HER2 copy number between plasma and tissue samples in gastric cancer using droplet digital PCR[J]. Sci Rep, 2020, 10(1): 4177. |
| 23 | WANG Y, TSANG J Y S, CUI Y, et al. Robust and accurate digital measurement for HER2 amplification in HER2 equivocal breast cancer diagnosis[J]. Sci Rep, 2017, 7(1): 6752. |
| 24 | WANG H, LI B, LIU Z, et al. HER2 copy number of circulating tumor DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer[J]. Eur J Cancer, 2018, 88:92-100. |
| 25 | LEE K S, NAM S K, SEO S H, et al. Digital polymerase chain reaction for detecting c-MYC copy number gain in tissue and cell-free plasma samples of colorectal cancer patients[J]. Sci Rep, 2019, 9(1): 1611. |
| [1] | 王静怡, 邓佳丽, 朱仪, 丁心怡, 郭嘉婧, 王中领. 新型pH响应性锰基纳米探针用于乳腺癌铁死亡及磁共振成像实验研究[J]. 上海交通大学学报(医学版), 2025, 45(9): 1183-1193. |
| [2] | 李龙, 赵霞, 金珊, 李泽莹, 吕福强, 庞丽娟, 刘克坚. 孟德尔随机化解析AZGP1在心力衰竭中的保护作用[J]. 上海交通大学学报(医学版), 2025, 45(8): 1035-1045. |
| [3] | 林桐, 陶怡, 金诗炜, 孙淼, 糜坚青. 血浆置换联合经典化学治疗对多发性骨髓瘤患者肾功能的影响[J]. 上海交通大学学报(医学版), 2025, 45(7): 823-828. |
| [4] | 宿星蕾, 路平, 彭俊杰, 汪滋民, 宋萍, 韩达. 血液样本中DNA与RNA提取效率的研究[J]. 上海交通大学学报(医学版), 2025, 45(4): 476-486. |
| [5] | 邓佳丽, 郭嘉婧, 王静怡, 丁心怡, 朱仪, 王中领. 自组装载药纳米探针用于乳腺癌焦亡增敏及化学交换饱和转移成像研究[J]. 上海交通大学学报(医学版), 2025, 45(3): 271-281. |
| [6] | 吴诗怡, 陈思, 刘泊含, 刘宇婷, 刘鷖雯, 何怡青, 杜艳, 张国良, 郭倩, 高锋, 杨翠霞. “HA糖外衣”调控ER+乳腺癌细胞干性在内分泌治疗耐药中的作用[J]. 上海交通大学学报(医学版), 2025, 45(10): 1298-1307. |
| [7] | 吴其蓁, 刘启明, 柴烨子, 陶政宇, 王依楠, 郭欣宁, 姜萌, 卜军. 机器学习预测乳腺癌新辅助治疗后炎症代谢状态改变的模型评价[J]. 上海交通大学学报(医学版), 2024, 44(9): 1169-1181. |
| [8] | 韩依杉, 徐梓淇, 陶梦玉, 范广建, 余波. PRMT6促进乳腺癌细胞的增殖和迁移[J]. 上海交通大学学报(医学版), 2024, 44(8): 999-1010. |
| [9] | 王卫, 王红丽, 阿力比亚提·艾尼, 衣力亚尔·肉苏, 阿依努尔, 杨亮. 血管抑制蛋白2在三阴性乳腺癌中的功能及其调控可变剪接机制[J]. 上海交通大学学报(医学版), 2024, 44(12): 1526-1535. |
| [10] | 谭辰, 徐张润, 薛阳, 陈佳钰, 姚力郡. 老药新用在乳腺癌治疗中的研究进展[J]. 上海交通大学学报(医学版), 2024, 44(11): 1454-1459. |
| [11] | 杜少倩, 陶梦玉, 曹源, 王红霞, 胡孝渠, 范广建, 臧丽娟. CXCL9在乳腺癌中的表达及其与肿瘤免疫浸润特征的相关性研究[J]. 上海交通大学学报(医学版), 2023, 43(7): 860-872. |
| [12] | 曹源, 王红霞, 朱灜, 李军建. 四次跨膜蛋白1在乳腺癌中的表达及其促进乳腺癌进展的作用机制[J]. 上海交通大学学报(医学版), 2023, 43(3): 293-300. |
| [13] | 杨笑萱, 朱珊, 钱程, 储晓英. 术中使用小剂量右美托咪定对乳腺癌手术患者预后的影响[J]. 上海交通大学学报(医学版), 2023, 43(2): 194-200. |
| [14] | 王斓茜, 马官荣, 姜咏竹, 常秀林, 方廖琼, 白晋. 大肠埃希菌外膜囊泡对乳腺癌细胞增殖及荷瘤小鼠肿瘤生长的影响[J]. 上海交通大学学报(医学版), 2023, 43(10): 1245-1254. |
| [15] | 夏坤健, 邓林林, 王琳. 乳腺癌化学治疗致肝损伤预测模型的构建及其评价[J]. 上海交通大学学报(医学版), 2022, 42(4): 502-509. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||